NLU Version 4.2.2
- support for Medical Summarizers
New Medical Summarizers:
- ‘en.summarize.clinical_jsl’
- ‘en.summarize.clinical_jsl_augmented’
- ‘en.summarize.biomedical_pubmed’
- ‘en.summarize.generic_jsl’
- ‘en.summarize.clinical_questions’
- ‘en.summarize.radiology’
- ‘en.summarize.clinical_guidelines_large’
- ‘en.summarize.clinical_laymen’
NLU Version 4.2.1
Bugfixes for saving and reloading pipelines on databricks
NLU Version 4.2.0
Support for Speech2Text, Images-Classification, Tabular Data, Zero-Shot-NER, via Wav2Vec2, Tapas, VIT , 4000+ New Models, 90+ Languages, in John Snow Labs NLU 4.2.0
We are incredibly excited to announce NLU 4.2.0 has been released with new 4000+ models in 90+ languages and support for new 8 Deep Learning Architectures. 4 new tasks are included for the very first time, Zero-Shot-NER, Automatic Speech Recognition, Image Classification and Table Question Answering powered by Wav2Vec 2.0, HuBERT, TAPAS, VIT, SWIN, Zero-Shot-NER.
Additionally, CamemBERT based architectures are available for Sequence and Token Classification powered by Spark-NLPs CamemBertForSequenceClassification and CamemBertForTokenClassification
Automatic Speech Recognition (ASR)
Demo Notebook Wav2Vec 2.0 and HuBERT enable ASR for the very first time in NLU. Wav2Vec2 is a transformer model for speech recognition that uses unsupervised pre-training on large amounts of unlabeled speech data to improve the accuracy of automatic speech recognition (ASR) systems. It is based on a self-supervised learning approach that learns to predict masked portions of speech signal, and has shown promising results in reducing the amount of labeled training data required for ASR tasks.
These Models are powered by Spark-NLP’s Wav2Vec2ForCTC Annotator

HuBERT models match or surpass the SOTA approaches for speech representation learning for speech recognition, generation, and compression. The Hidden-Unit BERT (HuBERT) approach was proposed for self-supervised speech representation learning, which utilizes an offline clustering step to provide aligned target labels for a BERT-like prediction loss.
These Models is powered by Spark-NLP’s HubertForCTC Annotator

Usage
You just need an audio-file on disk and pass the path to it or a folder of audio-files.
import nlu
# Let's download an audio file
!wget https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/audio/samples/wavs/ngm_12484_01067234848.wav
# Let's listen to it
from IPython.display import Audio
FILE_PATH = "ngm_12484_01067234848.wav"
asr_df = nlu.load('en.speech2text.wav2vec2.v2_base_960h').predict('ngm_12484_01067234848.wav')
asr_df
| text |
|---|
| PEOPLE WHO DIED WHILE LIVING IN OTHER PLACES |
To test out HuBERT you just need to update the parameter for load()
asr_df = nlu.load('en.speech2text.hubert').predict('ngm_12484_01067234848.wav')
asr_df
Image Classification
For the first time ever NLU introduces state-of-the-art image classifiers based on
VIT and Swin giving you access to hundreds of image classifiers for various domains.
Inspired by the Transformer scaling successes in NLP, the researchers experimented with applying a standard Transformer directly to images, with the fewest possible modifications. To do so, images are split into patches and the sequence of linear embeddings of these patches were provided as an input to a Transformer. Image patches were actually treated the same way as tokens (words) in an NLP application. Image classification models were trained in supervised fashion.
You can check Scale Vision Transformers (ViT) Beyond Hugging Face article to learn deeper how ViT works and how it is implemeted in Spark NLP. This is Powerd by Spark-NLP’s VitForImageClassification Annotator
Swin is a hierarchical Transformer whose representation is computed with Shifted windows. The shifted windowing scheme brings greater efficiency by limiting self-attention computation to non-overlapping local windows while also allowing for cross-window connection. This hierarchical architecture has the flexibility to model at various scales and has linear computational complexity with respect to image size. These qualities of Swin Transformer make it compatible with a broad range of vision tasks This is powerd by Spark-NLP’s Swin For Image Classification Swin Transformer: Hierarchical Vision Transformer using Shifted Windows by Ze Liu, Yutong Lin, Yue Cao, Han Hu, Yixuan Wei, Zheng Zhang, Stephen Lin, Baining Guo.

Usage:
# Download an image
os.system('wget https://raw.githubusercontent.com/JohnSnowLabs/nlu/release/4.2.0/tests/datasets/ocr/vit/ox.jpg')
# Load VIT model and predict on image file
vit = nlu.load('en.classify_image.base_patch16_224').predict('ox.jpg')
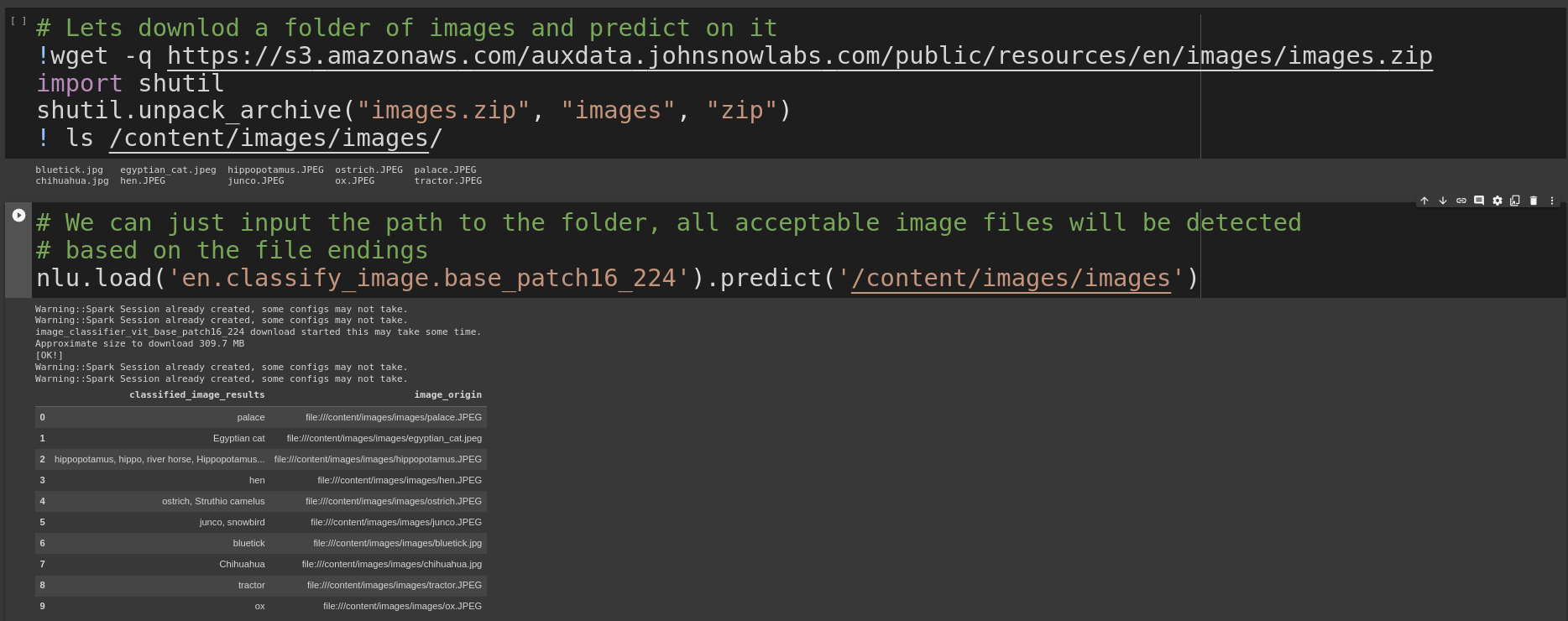
Lets download a folder of images and predict on it
!wget -q https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/images/images.zip
import shutil
shutil.unpack_archive("images.zip", "images", "zip")
! ls /content/images/images/
Once we have image data its easy to label it, we just pass the folder with images to nlu.predict() and NLU will return a pandas DF with one row per image detected
nlu.load('en.classify_image.base_patch16_224').predict('/content/images/images')
To use SWIN we just update the parameter to load()
load('en.classify_image.swin.tiny').predict('/content/images/images')
Visual Table Question Answering
TapasForQuestionAnswering can load TAPAS Models with a cell selection head and optional aggregation head on top for question-answering tasks on tables (linear layers on top of the hidden-states output to compute logits and optional logits_aggregation), e.g. for SQA, WTQ or WikiSQL-supervised tasks. TAPAS is a BERT-based model specifically designed (and pre-trained) for answering questions about tabular data.
Powered by TAPAS: Weakly Supervised Table Parsing via Pre-training

Usage:
First we need a pandas dataframe on for which we want to ask questions. The so called “context”
import pandas as pd
context_df = pd.DataFrame({
'name':['Donald Trump','Elon Musk'],
'money': ['$100,000,000','$20,000,000,000,000'],
'married': ['yes','no'],
'age' : ['75','55'] })
context_df
Then we create an array of questions
questions = [
"Who earns less than 200,000,000?",
"Who earns more than 200,000,000?",
"Who earns 100,000,000?",
"How much money has Donald Trump?",
"Who is the youngest?",
]
questions
Now Combine the data, pass it to NLU and get answers for your questions
import nlu
# Now we combine both to a tuple and we are done! We can now pass this to the .predict() method
tapas_data = (context_df, questions)
# Lets load a TAPAS QA model and predict on (context,question).
# It will give us an aswer for every question in the questions array, based on the context in context_df
answers = nlu.load('en.answer_question.tapas.wtq.large_finetuned').predict(tapas_data)
answers
| sentence | tapas_qa_UNIQUE_aggregation | tapas_qa_UNIQUE_answer | tapas_qa_UNIQUE_cell_positions | tapas_qa_UNIQUE_cell_scores | tapas_qa_UNIQUE_origin_question |
|---|---|---|---|---|---|
| Who earns less than 200,000,000? | NONE | Donald Trump | [0, 0] | 1 | Who earns less than 200,000,000? |
| Who earns more than 200,000,000? | NONE | Elon Musk | [0, 1] | 1 | Who earns more than 200,000,000? |
| Who earns 100,000,000? | NONE | Donald Trump | [0, 0] | 1 | Who earns 100,000,000? |
| How much money has Donald Trump? | SUM | SUM($100,000,000) | [1, 0] | 1 | How much money has Donald Trump? |
| Who is the youngest? | NONE | Elon Musk | [0, 1] | 1 | Who is the youngest? |
Zero-Shot NER
Demo Notebook
Based on John Snow Labs Enterprise-NLP ZeroShotNerModel
This architecture is based on RoBertaForQuestionAnswering.
Zero shot models excel at generalization, meaning that the model can accurately predict entities in very different data sets without the need to fine tune the model or train from scratch for each different domain.
Even though a model trained to solve a specific problem can achieve better accuracy than a zero-shot model in this specific task,
it probably won’t be be useful in a different task.
That is where zero-shot models shows its usefulness by being able to achieve good results in various domains.
Usage:
We just need to load the zero-shot NER model and configure a set of entity definitions.
import nlu
# load zero-shot ner model
enterprise_zero_shot_ner = nlu.load('en.zero_shot.ner_roberta')
# Configure entity definitions
enterprise_zero_shot_ner['zero_shot_ner'].setEntityDefinitions(
{
"PROBLEM": [
"What is the disease?",
"What is his symptom?",
"What is her disease?",
"What is his disease?",
"What is the problem?",
"What does a patient suffer",
"What was the reason that the patient is admitted to the clinic?",
],
"DRUG": [
"Which drug?",
"Which is the drug?",
"What is the drug?",
"Which drug does he use?",
"Which drug does she use?",
"Which drug do I use?",
"Which drug is prescribed for a symptom?",
],
"ADMISSION_DATE": ["When did patient admitted to a clinic?"],
"PATIENT_AGE": [
"How old is the patient?",
"What is the gae of the patient?",
],
}
)
Then we can already use this pipeline to predict labels
# Predict entities
df = enterprise_zero_shot_ner.predict(
[
"The doctor pescribed Majezik for my severe headache.",
"The patient was admitted to the hospital for his colon cancer.",
"27 years old patient was admitted to clinic on Sep 1st by Dr."+
"X for a right-sided pleural effusion for thoracentesis.",
]
)
df
| document | entities_zero_shot | entities_zero_shot_class | entities_zero_shot_confidence | entities_zero_shot_origin_chunk | entities_zero_shot_origin_sentence |
|---|---|---|---|---|---|
| The doctor pescribed Majezik for my severe headache. | Majezik | DRUG | 0.646716 | 0 | 0 |
| The doctor pescribed Majezik for my severe headache. | severe headache | PROBLEM | 0.552635 | 1 | 0 |
| The patient was admitted to the hospital for his colon cancer. | colon cancer | PROBLEM | 0.88985 | 0 | 0 |
| 27 years old patient was admitted to clinic on Sep 1st by Dr. X for a right-sided pleural effusion for thoracentesis. | 27 years old | PATIENT_AGE | 0.694308 | 0 | 0 |
| 27 years old patient was admitted to clinic on Sep 1st by Dr. X for a right-sided pleural effusion for thoracentesis. | Sep 1st | ADMISSION_DATE | 0.956461 | 1 | 0 |
| 27 years old patient was admitted to clinic on Sep 1st by Dr. X for a right-sided pleural effusion for thoracentesis. | a right-sided pleural effusion for thoracentesis | PROBLEM | 0.500266 | 2 | 0 |
New Notebooks
New Models Overview
Supported Languages are:
ab, am, ar, ba, bem, bg, bn, ca, co, cs, da, de, dv, el, en, es, et, eu, fa, fi, fon, fr, fy, ga, gam, gl, gu, ha, he, hi, hr, hu, id, ig, is, it, ja, jv, kin, kn, ko, kr, ku, ky, la, lg, lo, lt, lu, luo, lv, lwt, ml, mn, mr, ms, mt, nb, nl, no, pcm, pl, pt, ro, ru, rw, sg, si, sk, sl, sq, st, su, sv, sw, swa, ta, te, th, ti, tl, tn, tr, tt, tw, uk, unk, ur, uz, vi, wo, xx, yo, yue, zh, zu
Automatic Speech Recognition Models Overview
Image Classification Models Overview
NLU Version 4.1.0
Approximately 1000 new state-of-the-art transformer models for Question Answering (QA) for over 10 languages, up to 700% speedup on GPU, 100+ Embeddings such as Bert, Bert Sentence, CamemBert, DistilBert, Roberta, Roberta Sentence, Universal Sentence Encoder, Word, XLM Roberta, XLM Roberta Sentence, 40 sequence classification models, +400 token classification odels for over 10 languages various Spark NLP helper methods and much more in 1 line of code with John Snow Labs NLU 4.1.0
NLU 4.1.0 Core Overview
-
On the NLU core side we have over 1000 new state-of-the-art models in over 10 languages.
-
Additionally up to 700% speedup transformer-based Word Embeddings on GPU and up to 97% speedup on CPU for tensorflow operations, support for Apple M1 chips, Pyspark 3.2 and 3.3 support. Ontop of this, we are now supporting Apple M1 based architectures and every Pyspark 3.X version, while deprecating support for Pyspark 2.X.
-
Finally, NLU-Core features various new helper methods for working with Spark NLP and embellishes now the entire universe of Annotators defined by Spark NLP.
NLU captures every Annotator of Spark NLP
The entire universe of Annotators in Spark NLP is now embellished by NLU Components by using generalizable annotation extractors methods and configs internally to support enable the new NLU util methods. The following annotator classes are newly captured:
- BertEmbeddings
- BertForQuestionAnswering
- BertForSequenceClassification
- BertForTokenClassification
- BertSentenceEmbeddings
- CamemBertEmbeddings
- ClassifierDLModel
- ContextSpellCheckerModel
- DistilBertEmbeddings
- DistilBertForSequenceClassification
- DistilBertForTokenClassification
- LemmatizerModel
- LongformerForTokenClassification
- NerCrfModel
- NerDLModel
- PerceptronModel
- RoBertaEmbeddings
- RoBertaForQuestionAnswering
- RoBertaForSequenceClassification
- RoBertaForTokenClassification
- RoBertaSentenceEmbeddings
- SentenceDetectorDLModel
- StopWordsCleaner
- T5Transformer
- UniversalSentenceEncoder
- WordEmbeddingsModel
- XlmRoBertaEmbeddings
- XlmRoBertaForTokenClassification
- XlmRoBertaSentenceEmbeddings
Embeddings
Embeddings provides dense vector representations for natural language by using a deep, pre-trained neural network with the Transformer architecture. On the NLU core side we have over 150 new embeddings models. We have new BertEmbeddings, BertSentenceEmbeddings, CamemBertEmbeddings, DistilBertEmbeddings, RoBertaEmbeddings, UniversalSentenceEncoder, XlmRoBertaEmbeddings, XlmRoBertaSentenceEmbeddings for in different languages.
- German BertEmbeddings
nlu.load("de.embed.electra.base").predict("""Ich liebe Spark NLP""")
| token | word_embedding_electra |
|---|---|
| Ich | -0.09518987685441971, -0.016133345663547516 |
| liebe | -0.07025116682052612, -0.35387516021728516 |
| Spark | -0.33390265703201294, 0.08874476701021194 |
| NLP | -0.2969835698604584, 0.1980721354484558 |
- English BertEmbeddings
text = ["I love NLP"]
df = nlu.load('en.embed_sentence.bert.pubmed').predict(text, output_level='token')
df
| token | sentence_embedding_bert |
|---|---|
| I | -0.06332794576883316, -0.5097940564155579 |
| love | -0.06332794576883316, -0.5097940564155579 |
| NLP | -0.06332794576883316, -0.5097940564155579 |
- Japan BertEmbeddings
nlu.load("ja.embed.bert.base").predict("""私はSpark NLPを愛しています""")
| token | word_embedding_bert |
|---|---|
| 私はSpark | 0.3989057242870331, -0.20664098858833313 |
| NLPを愛しています | 0.05264343321323395, -0.19963961839675903 |
- XLM RoBerta Embeddings MultiLanguage
text = ["I love NLP", "Me encanta usar SparkNLP"]
embeddings_df = nlu.load('xx.embed.xlmr_roberta.base_v2').predict(text, output_level='sentence')
embeddings_df
| sentence | word_embedding_xlmr_roberta |
|---|---|
| I love NLP | -0.07450243085622787, 0.022609828040003777 |
| Me encanta usar SparkNLP | 0.0961054190993309, 0.03734250366687775 |
- RoBerta Embeddings English
text = ["""I love Spark NLP"""]
embeddings_df = nlu.load('en.embed.roberta').predict(text, output_level='token')
embeddings_df
| token | word_embedding_roberta |
|---|---|
| I | -0.06406927853822708, 0.16723069548606873 |
| love | -0.06369957327842712, 0.21014901995658875 |
| Spark | -0.1004200279712677, 0.03312099352478981 |
| NLP | -0.09467814117670059, -0.02236207202076912 |
Question Answering
Question Answering models can retrieve the answer to a question from a given text, which is useful for searching for an answer in a document. On the NLU core side we have over 200+ new question answering models.
- Bert For Question Answering
nlu.load("answer_question.bert.base_uncased.by_ksabeh").predict("""What is my name?|||"My name is Clara and I live in Berkeley.""")
| answer_confidence | context | question |
|---|---|---|
| 0.3143375 | “My name is Clara and I live in Berkeley. | What is my name? |
Sequence Classification
Sequence classification is the task of predicting a class label given a sequence of observations. On the NLU core side we have over 40 new sequence classification models.
- Bert For Sequence Classification
nlu.load("classify.bert.by_mrm8488").predict("""Camera - You are awarded a SiPix Digital Camera! call 09061221066 from landline. Delivery within 28 days.""")
| classified_sequence | classified_sequence_confidence | sentence |
|---|---|---|
| 1 | 0.89954 | Camera - You are awarded a SiPix Digital Camera! call 09061221066 from landline. |
| 0 | 0.93745 | Delivery within 28 days. |
- DistilBert For Sequence Classification
nlu.load("de.classify.distil_bert.base").predict("Natürlich kann ich von zuwanderern mehr erwarten. muss ich sogar. sie müssen die sprache lernen, sie müssen die gepflogenheiten lernen und sich in die gesellschaft einfügen. dass muss ich nicht weil ich mich schon in die gesellschaft eingefügt habe. egal wo du hin ziehst, nirgendwo wird dir soviel zucker in den arsch geblasen wie in deutschland.")
| classified_sequence | classified_sequence_confidence | sentence |
|---|---|---|
| non_toxic | 0.955292 | Natürlich kann ich von zuwanderern mehr erwarten. |
| non_toxic | 0.968591 | muss ich sogar. |
| non_toxic | 0.841958 | sie müssen die sprache lernen, sie müssen die gepflogenheiten lernen und sich in die gesellschaft einfügen. |
| non_toxic | 0.934119 | dass muss ich nicht weil ich mich schon in die gesellschaft eingefügt habe. |
| non_toxic | 0.771795 | egal wo du hin ziehst, nirgendwo wird dir soviel zucker in den arsch geblasen wie in deutschland. |
- RoBerta For Sequence Classification
nlu.load("en.classify.roberta.finetuned").predict("I love you very much!")
| classified_sequence | classified_sequence_confidence | sentence |
|---|---|---|
| LABEL_0 | 0.597792 | I love you very much! |
Lemmatizer
Lemmatization in linguistics is the process of grouping together the inflected forms of a word so they can be analysed as a single item, identified by the word’s lemma, or dictionary form. On the NLU core side we have over 30 new lemmatizer models.
ClassifierDLModel
ClassifierDL for generic Multi-class Text Classification. ClassifierDL uses the state-of-the-art Universal Sentence Encoder as an input for text classifications. The ClassifierDL annotator uses a deep learning model (DNNs) we have built inside TensorFlow and supports up to 100 classes. On the NLU core side we have over 5 new ClassifierDLModel models.
ContextSpellCheckerModel
Spell Checking is a sequence to sequence mapping problem. Given an input sequence, potentially containing a certain number of errors, ContextSpellChecker will rank correction sequences according to three things:
- Different correction candidates for each word — word level.
- The surrounding text of each word, i.e. it’s context — sentence level.
- The relative cost of different correction candidates according to the edit operations at the character level it requires — subword level.
On the NLU core side we have over 5 new ClassifierDLModel models.
Token Classification
Token classification is a natural language understanding task in which a label is assigned to some tokens in a text. Some popular token classification subtasks are Named Entity Recognition (NER) and Part-of-Speech (PoS) tagging. NER models could be trained to identify specific entities in a text, such as dates, individuals and places; and PoS tagging would identify, for example, which words in a text are verbs, nouns, and punctuation marks. We have new 463 models XlmRoBertaForTokenClassification, BertForTokenClassification, DistilBertForTokenClassification, DistilBertEmbeddings, LongformerForTokenClassification, RoBertaForTokenClassification for in different languages.
- BertForTokenClassification English
nlu.load("en.ner.bc5cdr.biobert.disease").predict("I love you very much!")
| index | document | entities_wikiner_glove_840B_300 | entities_wikiner_glove_840B_300_class | entities_wikiner_glove_840B_300_confidence | entities_wikiner_glove_840B_300_origin_chunk | entities_wikiner_glove_840B_300_origin_sentence | word_embedding_glove |
|---|---|---|---|---|---|---|---|
| 0 | I love you very much! | I love you very much! | MISC | 0.66433334 | 0 | 0 | [ 0.19410001 0.22603001 -0.43764001 ] |
- BertForTokenClassification German
nlu.load("de.ner.distil_bert.base_cased").predict("Ich liebe Spark NLP")
| index | classified_token | document | entities_distil_bert | entities_distil_bert_class | entities_distil_bert_origin_chunk | entities_distil_bert_origin_sentence |
|---|---|---|---|---|---|---|
| 0 | O,O,B-OTHderiv,O | Ich liebe Spark NLP | Spark | OTHderiv | 0 | 0 |
- XlmRoBertaForTokenClassification Igbo
nlu.load("ig.ner.xlmr_roberta.base").predict("Ahụrụ m n'anya na-atọ m ụtọ")
| index | classified_token | document | entities_xlmr_roberta | entities_xlmr_roberta_class | entities_xlmr_roberta_origin_chunk | entities_xlmr_roberta_origin_sentence |
|---|---|---|---|---|---|---|
| 0 | B-ORG,I-ORG,I-ORG,I-ORG,I-ORG,I-ORG | Ahụrụ m n’anya na-atọ m ụtọ | Ahụrụ m n’anya na-atọ m ụtọ | ORG | 0 | 0 |
NerCrfModel
This Named Entity Recognizer is based on a CRF Algorithm. Conditional random fields (CRFs) are a class of statistical modeling methods often applied in pattern recognition and machine learning and used for structured prediction. Whereas a classifier predicts a label for a single sample without considering “neighbouring” samples, a CRF can take context into account. To do so, the predictions are modelled as a graphical model, which represents the presence of dependencies between the predictions. What kind of graph is used depends on the application. For example, in natural language processing, “linear chain” CRFs are popular, for which each prediction is dependent only on its immediate neighbours. In image processing, the graph typically connects locations to nearby and/or similar locations to enforce that they receive similar predictions.
- NerCrfModel
nlu.load('en.ner.ner.crf').predict("Donald Trump and Angela Merkel dont share many oppinions")
| index | document | entities_wikiner_glove_840B_300 | entities_wikiner_glove_840B_300_class | entities_wikiner_glove_840B_300_confidence | entities_wikiner_glove_840B_300_origin_chunk | entities_wikiner_glove_840B_300_origin_sentence | word_embedding_glove |
|---|---|---|---|---|---|---|---|
| 0 | Donald Trump and Angela Merkel dont share many oppinions | Donald Trump | PER | 0.78524995 | 0 | 0 | [-0.074014 -0.23684999 0.17772 ] |
| 0 | Donald Trump and Angela Merkel dont share many oppinions | Angela Merkel | PER | 0.7701 | 1 | 0 | [-0.074014 -0.23684999 0.17772 ] |
NerDLModel
This Named Entity recognition annotator is a generic NER model based on Neural Networks. Neural Network architecture is Char CNNs - BiLSTM - CRF that achieves state-of-the-art in most datasets. This is the instantiated model of the NerDLApproach. For training your own model, please see the documentation of that class. We have new 6 models.
- NerDLModel Japanese
nlu.load('ja.ner.ner.base').predict("宮本茂氏は、日本の任天堂のゲームプロデューサーです。")
| index | document | entities_xtreme_glove_840B_300 | word_embedding_glove |
|---|---|---|---|
| 0 | 宮本茂氏は、日本の任天堂のゲームプロデューサーです。 | NaN | [0. 0. ] |
- NerDLModel English
text = ["My name is John!"]
nlu.load('en.ner.conll.ner.large').predict(text, output_level='token')
| index | entities_wikiner_glove_840B_300 | entities_wikiner_glove_840B_300_class | entities_wikiner_glove_840B_300_confidence | entities_wikiner_glove_840B_300_origin_chunk | entities_wikiner_glove_840B_300_origin_sentence | token | word_embedding_glove |
|---|---|---|---|---|---|---|---|
| 0 | My name is John! | MISC | 0.63266003 | 0 | 0 | My | [-2.19990000e-01 2.57800013e-01 -4.25859988e-01 ] |
| 0 | My name is John! | MISC | 0.63266003 | 0 | 0 | name | [ 2.32309997e-01 -2.41020005e-02] |
| 0 | My name is John! | MISC | 0.63266003 | 0 | 0 | is | [-8.49609971e-02 5.01999974e-01 2.38230010e-03] |
| 0 | My name is John! | MISC | 0.63266003 | 0 | 0 | John | [-2.96090007e-01 -8.18260014e-02 9.67490021e-03 ] |
| 0 | My name is John! | MISC | 0.63266003 | 0 | 0 | ! | [-2.65540004e-01 3.35310012e-01 2.18600005e-01 ] |
PerceptronModel
We have new 26 models.
StopWordsCleaner
This model removes ‘stop words’ from text. Stop words are words so common that they can be removed without significantly altering the meaning of a text. Removing stop words is useful when one wants to deal with only the most semantically important words in a text, and ignore words that are rarely semantically relevant, such as articles and prepositions. We have new 33 models.
NLU Version 4.0.0
OCR Visual Tables into Pandas DataFrames from PDF/DOC(X)/PPT files, 1000+ new state-of-the-art transformer models for Question Answering (QA) for over 30 languages, up to 700% speedup on GPU, 20 Biomedical models for over 8 languages, 50+ Terminology Code Mappers between RXNORM, NDC, UMLS,ICD10, ICDO, UMLS, SNOMED and MESH, Deidentification in Romanian, various Spark NLP helper methods and much more in 1 line of code with John Snow Labs NLU 4.0.0
NLU 4.0 for OCR Overview
On the OCR side, we now support extracting tables from PDF/DOC(X)/PPT files into structured pandas dataframe, making it easier than ever before to analyze bulks of files visually!
Checkout the OCR Tutorial for extracting Tables from Image/PDF/DOC(X) files to see this in action
These models grab all Table data from the files detected and return a list of Pandas DataFrames,
containing Pandas DataFrame for every table detected
| NLU Spell | Transformer Class |
|---|---|
nlu.load(pdf2table) |
PdfToTextTable |
nlu.load(ppt2table) |
PptToTextTable |
nlu.load(doc2table) |
DocToTextTable |
This is powerd by John Snow Labs Spark OCR Annotataors for PdfToTextTable, DocToTextTable, PptToTextTable
NLU 4.0 Core Overview
-
On the NLU core side we have over 1000+ new state-of-the-art models in over 30 languages for modern extractive transformer-based Question Answering problems powerd by the ALBERT/BERT/DistilBERT/DeBERTa/RoBERTa/Longformer Spark NLP Annotators trained on various SQUAD-like QA datasets for domains like Twitter, Tech, News, Biomedical COVID-19 and in various model subflavors like sci_bert, electra, mini_lm, covid_bert, bio_bert, indo_bert, muril, sapbert, bioformer, link_bert, mac_bert
-
Additionally up to 700% speedup transformer-based Word Embeddings on GPU and up to 97% speedup on CPU for tensorflow operations, support for Apple M1 chips, Pyspark 3.2 and 3.3 support. Ontop of this, we are now supporting Apple M1 based architectures and every Pyspark 3.X version, while deprecating support for Pyspark 2.X.
-
Finally, NLU-Core features various new helper methods for working with Spark NLP and embellishes now the entire universe of Annotators defined by Spark NLP and Spark NLP for healthcare.
NLU 4.0 for Healthcare Overview
-
On the healthcare side NLU features 20 Biomedical models for over 8 languages (English, French, Italian, Portuguese, Romanian, Catalan and Galician) detect entities like
HUMANandSPECIESbased on LivingNER corpus -
Romanian models for Deidentification and extracting Medical entities like
Measurements,Form,Symptom,Route,Procedure,Disease_Syndrome_Disorder,Score,Drug_Ingredient,Pulse,Frequency,Date,Body_Part,Drug_Brand_Name,Time,Direction,Dosage,Medical_Device,Imaging_Technique,Test,Imaging_Findings,Imaging_Test,Test_Result,Weight,Clinical_DeptandUnitswith SPELL and SPELL respectively -
English NER models for parsing entities in Clinical Trial Abstracts like
Age,AllocationRatio,Author,BioAndMedicalUnit,CTAnalysisApproach,CTDesign,Confidence,Country,DisorderOrSyndrome,DoseValue,Drug,DrugTime,Duration,Journal,NumberPatients,PMID,PValue,PercentagePatients,PublicationYear,TimePoint,Valueusingen.med_ner.clinical_trials_abstracts.pipeand also Pathogen NER models forPathogen,MedicalCondition,Medicinewithen.med_ner.pathogenandGENE_PROTEINwithen.med_ner.biomedical_bc2gm.pipeline -
First Public Health Model for Emotional Stress classification It is a PHS-BERT-based model and trained with the Dreaddit dataset using
en.classify.stress -
50 + new Entity Mappers for problems like :
- Extract section headers in scientific articles and normalize them with
en.map_entity.section_headers_normalized - Map medical abbreviates to their definitions with
en.map_entity.abbreviation_to_definition - Map drugs to action and treatments with
en.map_entity.drug_to_action_treatment - Map drug brand to their National Drug Code (NDC) with
en.map_entity.drug_brand_to_ndc - Convert between terminologies using
en.<START_TERMINOLOGY>_to_<TARGET_TERMINOLOGY>- This works for the terminologies
rxnorm,ndc,umls,icd10cm,icdo,umls,snomed,meshsnomed_to_icdosnomed_to_icd10cmrxnorm_to_umls
- This works for the terminologies
- powerd by Spark NLP for Healthcares ChunkMapper Annotator
- Extract section headers in scientific articles and normalize them with
Extract Tables from PDF files as Pandas DataFrames
Sample PDF:
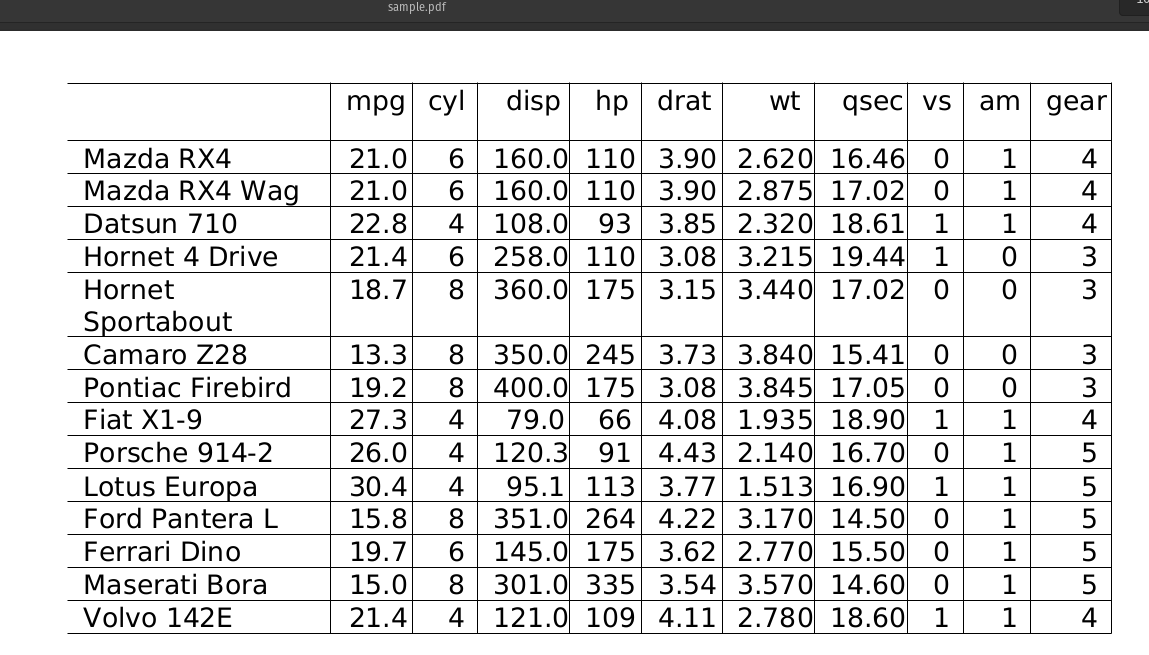
nlu.load('pdf2table').predict('/path/to/sample.pdf')
Output of PDF Table OCR :
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear |
|---|---|---|---|---|---|---|---|---|---|
| 21 | 6 | 160 | 110 | 3.9 | 2.62 | 16.46 | 0 | 1 | 4 |
| 21 | 6 | 160 | 110 | 3.9 | 2.875 | 17.02 | 0 | 1 | 4 |
| 22.8 | 4 | 108 | 93 | 3.85 | 2.32 | 18.61 | 1 | 1 | 4 |
| 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 |
| 18.7 | 8 | 360 | 175 | 3.15 | 3.44 | 17.02 | 0 | 0 | 3 |
| 13.3 | 8 | 350 | 245 | 3.73 | 3.84 | 15.41 | 0 | 0 | 3 |
| 19.2 | 8 | 400 | 175 | 3.08 | 3.845 | 17.05 | 0 | 0 | 3 |
| 27.3 | 4 | 79 | 66 | 4.08 | 1.935 | 18.9 | 1 | 1 | 4 |
| 26 | 4 | 120.3 | 91 | 4.43 | 2.14 | 16.7 | 0 | 1 | 5 |
| 30.4 | 4 | 95.1 | 113 | 3.77 | 1.513 | 16.9 | 1 | 1 | 5 |
| 15.8 | 8 | 351 | 264 | 4.22 | 3.17 | 14.5 | 0 | 1 | 5 |
| 19.7 | 6 | 145 | 175 | 3.62 | 2.77 | 15.5 | 0 | 1 | 5 |
| 15 | 8 | 301 | 335 | 3.54 | 3.57 | 14.6 | 0 | 1 | 5 |
| 21.4 | 4 | 121 | 109 | 4.11 | 2.78 | 18.6 | 1 | 1 | 4 |
Extract Tables from DOC/DOCX files as Pandas DataFrames
Sample DOCX:
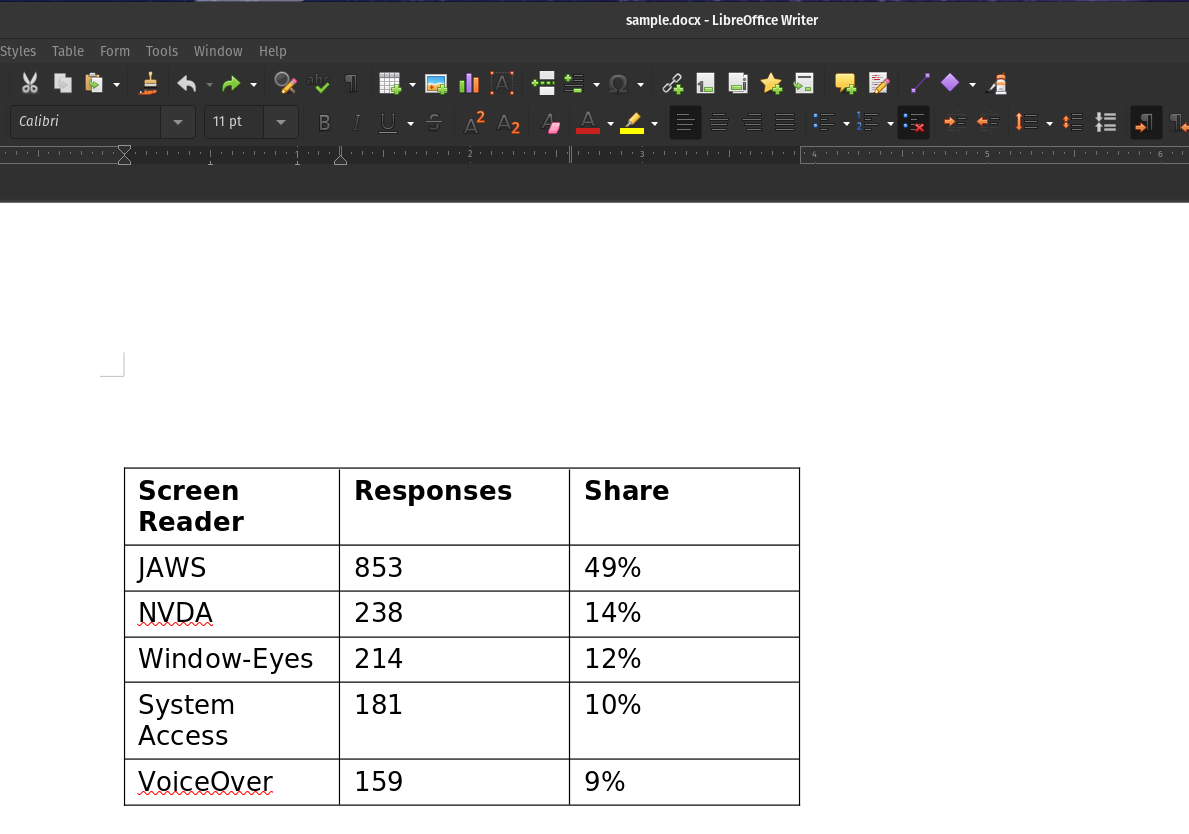
nlu.load('doc2table').predict('/path/to/sample.docx')
Output of DOCX Table OCR :
| Screen Reader | Responses | Share |
|---|---|---|
| JAWS | 853 | 49% |
| NVDA | 238 | 14% |
| Window-Eyes | 214 | 12% |
| System Access | 181 | 10% |
| VoiceOver | 159 | 9% |
Extract Tables from PPT files as Pandas DataFrame
Sample PPT with two tables:

nlu.load('ppt2table').predict('/path/to/sample.docx')
Output of PPT Table OCR :
{:.table-model-big}{:.table-model-big}
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|—————:|————–:|—————:|————–:|:———-|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
and
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 6.7 | 3.3 | 5.7 | 2.5 | virginica |
| 6.7 | 3 | 5.2 | 2.3 | virginica |
| 6.3 | 2.5 | 5 | 1.9 | virginica |
| 6.5 | 3 | 5.2 | 2 | virginica |
| 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 5.9 | 3 | 5.1 | 1.8 | virginica |
Span Classifiers for question answering
Albert, Bert, DeBerta, DistilBert, LongFormer, RoBerta, XlmRoBerta based Transformer Architectures are now avaiable for question answering with almost 1000 models avaiable for 35 unique languages powerd by their corrosponding Spark NLP XXXForQuestionAnswering Annotator Classes and in various tuning and dataset flavours.
<lang>.answer_question.<domain>.<datasets>.<annotator_class><tune info>.by_<username>
If multiple datasets or tune parameters are defined , they are connected with a _ .
These substrings define up the <domain> part of the NLU reference
- Legal cuad
- COVID 19 Biomedical biosaq
- Biomedical Literature pubmed
- Twitter tweet
- Wikipedia wiki
- News news
- Tech tech
These substrings define up the <dataset> part of the NLU reference
- Arabic SQUAD ARCD
- Turkish TQUAD
- German GermanQuad
- Indonesian AQG
- Korean KLUE, KORQUAD
- HindiCHAI
- Multi-LingualMLQA
- Multi-Lingualtydiqa
- Multi-Lingualxquad
These substrings define up the <dataset> part of the NLU reference
- Alternative Eval method reqa
- Synthetic Data synqa
- Benchmark / Eval Method ABSA-Bench roberta_absa
- Arabic architecture type soqaol
These substrings define the <annotator_class> substring, if it does not map to a sparknlp annotator
These substrings define the <tune_info> substring, if it does not map to a sparknlp annotator
- Train tweaks :
multilingual,mini_lm,xtremedistiled,distilled,xtreme,augmented,zero_shot - Size tweaks
xl,xxl,large,base,medium,base,small,tiny,cased,uncased - Dimension tweaks :
1024d,768d,512d,256d,128d,64d,32d
QA DataFormat
You need to use one of the Data formats below to pass context and question correctly to the model.
# use ||| to seperate question||context
data = 'What is my name?|||My name is Clara and I live in Berkeley'
# pass a tuple (question,context)
data = ('What is my name?','My name is Clara and I live in Berkeley')
# use pandas Dataframe, one column = question, one column=context
data = pd.DataFrame({
'question': ['What is my name?'],
'context': ["My name is Clara and I live in Berkely"]
})
# Get your answers with any of above formats
nlu.load("en.answer_question.squadv2.deberta").predict(data)
returns :
| answer | answer_confidence | context | question |
|---|---|---|---|
| Clara | 0.994931 | My name is Clara and I live in Berkely | What is my name? |
New NLU helper Methods
You can see all features showcased in the notebook or on the new docs page for Spark NLP utils
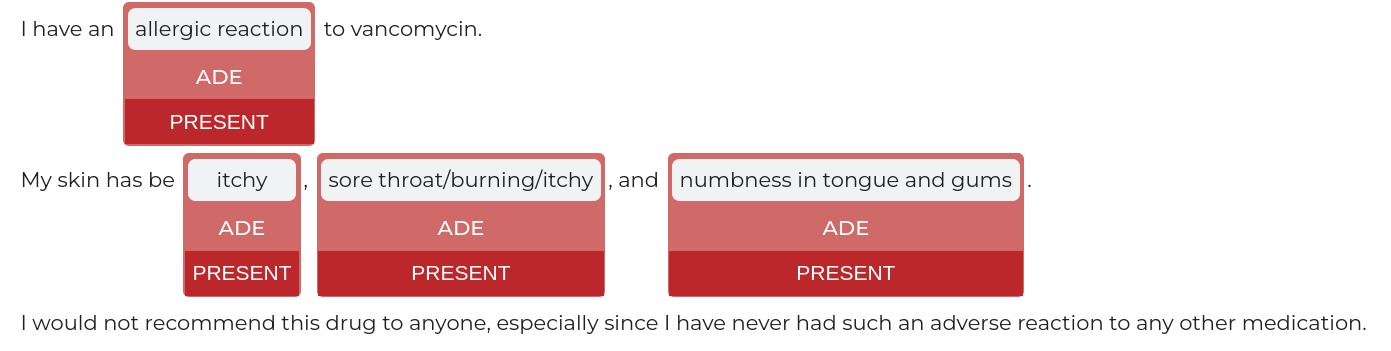
nlu.viz(pipe,data)
Visualize input data with an already configured Spark NLP pipeline,
for Algorithms of type (Ner,Assertion, Relation, Resolution, Dependency)
using Spark NLP Display
Automatically infers applicable viz type and output columns to use for visualization.
Example:
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlu.viz(ade_pipeline, text)
returns:
If a pipeline has multiple models candidates that can be used for a viz,
the first Annotator that is vizzable will be used to create viz.
You can specify which type of viz to create with the viz_type parameter
Output columns to use for the viz are automatically deducted from the pipeline, by using the
first annotator that provides the correct output type for a specific viz.
You can specify which columns to use for a viz by using the
corresponding ner_col, pos_col, dep_untyped_col, dep_typed_col, resolution_col, relation_col, assertion_col, parameters.
nlu.autocomplete_pipeline(pipe)
Auto-Complete a pipeline or single annotator into a runnable pipeline by harnessing NLU’s DAG Autocompletion algorithm and returns it as NLU pipeline.
The standard Spark pipeline is avaiable on the .vanilla_transformer_pipe attribute of the returned nlu pipe
Every Annotator and Pipeline of Annotators defines a DAG of tasks, with various dependencies that must be satisfied in topoligical order.
NLU enables the completion of an incomplete DAG by finding or creating a path between
the very first input node which is almost always is DocumentAssembler/MultiDocumentAssembler
and the very last node(s), which is given by the topoligical sorting the iterable annotators parameter.
Paths are created by resolving input features of annotators to the corrrosponding providers with matching storage references.
Example:
# Lets autocomplete the pipeline for a RelationExtractionModel, which as many input columns and sub-dependencies.
from sparknlp_jsl.annotator import RelationExtractionModel
re_model = RelationExtractionModel().pretrained("re_ade_clinical", "en", 'clinical/models').setOutputCol('relation')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlu_pipe = nlu.autocomplete_pipeline(re_model)
nlu_pipe.predict(text)
returns :
| relation | relation_confidence | relation_entity1 | relation_entity2 | relation_entity2_class |
|---|---|---|---|---|
| 1 | 1 | allergic reaction | vancomycin | Drug_Ingredient |
| 1 | 1 | skin | itchy | Symptom |
| 1 | 0.99998 | skin | sore throat/burning/itchy | Symptom |
| 1 | 0.956225 | skin | numbness | Symptom |
| 1 | 0.999092 | skin | tongue | External_body_part_or_region |
| 0 | 0.942927 | skin | gums | External_body_part_or_region |
| 1 | 0.806327 | itchy | sore throat/burning/itchy | Symptom |
| 1 | 0.526163 | itchy | numbness | Symptom |
| 1 | 0.999947 | itchy | tongue | External_body_part_or_region |
| 0 | 0.994618 | itchy | gums | External_body_part_or_region |
| 0 | 0.994162 | sore throat/burning/itchy | numbness | Symptom |
| 1 | 0.989304 | sore throat/burning/itchy | tongue | External_body_part_or_region |
| 0 | 0.999969 | sore throat/burning/itchy | gums | External_body_part_or_region |
| 1 | 1 | numbness | tongue | External_body_part_or_region |
| 1 | 1 | numbness | gums | External_body_part_or_region |
| 1 | 1 | tongue | gums | External_body_part_or_region |
nlu.to_pretty_df(pipe,data)
Annotates a Pandas Dataframe/Pandas Series/Numpy Array/Spark DataFrame/Python List strings /Python String
with given Spark NLP pipeline, which is assumed to be complete and runnable and returns it in a pythonic pandas dataframe format.
Example:
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
# output is same as nlu.autocomplete_pipeline(re_model).nlu_pipe.predict(text)
nlu.to_pretty_df(ade_pipeline,text)
returns :
| assertion | asserted_entitiy | entitiy_class | assertion_confidence |
|---|---|---|---|
| present | allergic reaction | ADE | 0.998 |
| present | itchy | ADE | 0.8414 |
| present | sore throat/burning/itchy | ADE | 0.9019 |
| present | numbness in tongue and gums | ADE | 0.9991 |
Annotators are grouped internally by NLU into output levels token,sentence, document,chunk and relation
Same level annotators output columns are zipped and exploded together to create the final output df.
Additionally, most keys from the metadata dictionary in the result annotations will be collected and expanded into their own columns in the resulting Dataframe, with special handling for Annotators that encode multiple metadata fields inside of one, seperated by strings like ||| or :::.
Some columns are omitted from metadata to reduce total amount of output columns, these can be re-enabled by setting metadata=True
For a given pipeline output level is automatically set to the last anntators output level by default.
This can be changed by defining to_preddty_df(pipe,text,output_level='my_level' for levels token,sentence, document,chunk and relation .
nlu.to_nlu_pipe(pipe)
Convert a pipeline or list of annotators into a NLU pipeline making .predict() and .viz() avaiable for every Spark NLP pipeline.
Assumes the pipeline is already runnable.
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlu_pipe = nlu.to_nlu_pipe(ade_pipeline)
# Same output as nlu.to_pretty_df(pipe,text)
nlu_pipe.predict(text)
# same output as nlu.viz(pipe,text)
nlu_pipe.viz(text)
# Acces auto-completed Spark NLP big data pipeline,
nlu_pipe.vanilla_transformer_pipe.transform(spark_df)
returns :
| assertion | asserted_entitiy | entitiy_class | assertion_confidence |
|---|---|---|---|
| present | allergic reaction | ADE | 0.998 |
| present | itchy | ADE | 0.8414 |
| present | sore throat/burning/itchy | ADE | 0.9019 |
| present | numbness in tongue and gums | ADE | 0.9991 |
and

4 new Demo Notebooks
These notebooks showcase some of latest classifier models for Banking Queries, Intents in Text, Question and new s classification
- Notebook for Classification of Banking Queries
- Notebook for Classification of Intent in Texts
- Notebook for classification of Similar Questions
- Notebook for Classification of Questions vs Statements
- Notebook for Classification of News into 4 classes
NLU captures every Annotator of Spark NLP and Spark NLP for healthcare
The entire universe of Annotators in Spark NLP and Spark-NLP for healthcare is now embellished by NLU Components by using generalizable annotation extractors methods and configs internally to support enable the new NLU util methods. The following annotator classes are newly captured:
- AssertionFilterer
- ChunkConverter
- ChunkKeyPhraseExtraction
- ChunkSentenceSplitter
- ChunkFiltererApproach
- ChunkFilterer
- ChunkMapperApproach
- ChunkMapperFilterer
- DocumentLogRegClassifierApproach
- DocumentLogRegClassifierModel
- ContextualParserApproach
- ReIdentification
- NerDisambiguator
- NerDisambiguatorModel
- AverageEmbeddings
- EntityChunkEmbeddings
- ChunkMergeApproach
- ChunkMergeApproach
- IOBTagger
- NerChunker
- NerConverterInternalModel
- DateNormalizer
- PosologyREModel
- RENerChunksFilter
- ResolverMerger
- AnnotationMerger
- Router
- Word2VecApproach
- WordEmbeddings
- EntityRulerApproach
- EntityRulerModel
- TextMatcherModel
- BigTextMatcher
- BigTextMatcherModel
- DateMatcher
- MultiDateMatcher
- RegexMatcher
- TextMatcher
- NerApproach
- NerCrfApproach
- NerOverwriter
- DependencyParserApproach
- TypedDependencyParserApproach
- SentenceDetectorDLApproach
- SentimentDetector
- ViveknSentimentApproach
- ContextSpellCheckerApproach
- NorvigSweetingApproach
- SymmetricDeleteApproach
- ChunkTokenizer
- ChunkTokenizerModel
- RecursiveTokenizer
- RecursiveTokenizerModel
- Token2Chunk
- WordSegmenterApproach
- GraphExtraction
- Lemmatizer
- Normalizer
All NLU 4.0 for Healthcare Models
Some examples:
en.rxnorm.umls.mapping
Code:
nlu.load('en.rxnorm.umls.mapping').predict('1161611 315677')
| mapped_entity_umls_code_origin_entity | mapped_entity_umls_code |
|---|---|
| 1161611 | C3215948 |
| 315677 | C0984912 |
en.ner.clinical_trials_abstracts
Code:
nlu.load('en.ner.clinical_trials_abstracts').predict('A one-year, randomised, multicentre trial comparing insulin glargine with NPH insulin in combination with oral agents in patients with type 2 diabetes.')
Results:
| entities_clinical_trials_abstracts | entities_clinical_trials_abstracts_class | entities_clinical_trials_abstracts_confidence | |
|---|---|---|---|
| 0 | randomised | CTDesign | 0.9996 |
| 0 | multicentre | CTDesign | 0.9998 |
| 0 | insulin glargine | Drug | 0.99135 |
| 0 | NPH insulin | Drug | 0.96875 |
| 0 | type 2 diabetes | DisorderOrSyndrome | 0.999933 |
Code:
nlu.load('en.ner.clinical_trials_abstracts').viz('A one-year, randomised, multicentre trial comparing insulin glargine with NPH insulin in combination with oral agents in patients with type 2 diabetes.')
Results:

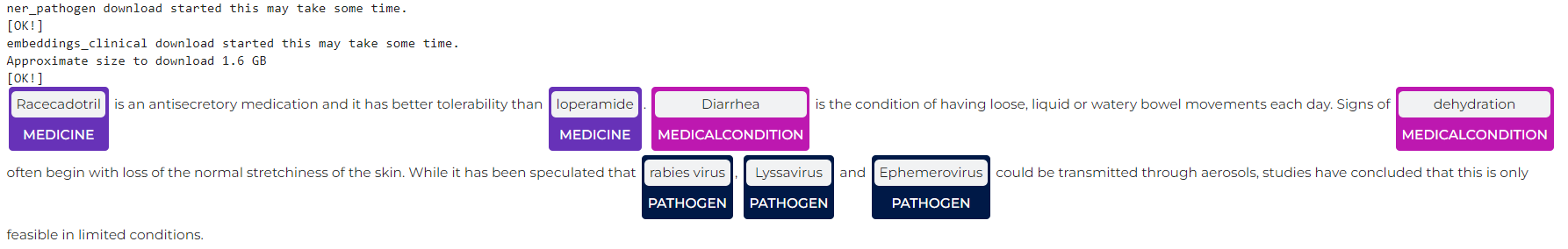
en.med_ner.pathogen
Code:
nlu.load('en.med_ner.pathogen').predict('Racecadotril is an antisecretory medication and it has better tolerability than loperamide. Diarrhea is the condition of having loose, liquid or watery bowel movements each day. Signs of dehydration often begin with loss of the normal stretchiness of the skin. While it has been speculated that rabies virus, Lyssavirus and Ephemerovirus could be transmitted through aerosols, studies have concluded that this is only feasible in limited conditions.')
Results:
| entities_pathogen | entities_pathogen_class | entities_pathogen_confidence | |
|---|---|---|---|
| 0 | Racecadotril | Medicine | 0.9468 |
| 0 | loperamide | Medicine | 0.9987 |
| 0 | Diarrhea | MedicalCondition | 0.9848 |
| 0 | dehydration | MedicalCondition | 0.6307 |
| 0 | rabies virus | Pathogen | 0.95685 |
| 0 | Lyssavirus | Pathogen | 0.9694 |
| 0 | Ephemerovirus | Pathogen | 0.6917 |
Code:
nlu.load('en.med_ner.pathogen').viz('Racecadotril is an antisecretory medication and it has better tolerability than loperamide. Diarrhea is the condition of having loose, liquid or watery bowel movements each day. Signs of dehydration often begin with loss of the normal stretchiness of the skin. While it has been speculated that rabies virus, Lyssavirus and Ephemerovirus could be transmitted through aerosols, studies have concluded that this is only feasible in limited conditions.')
Results:
es.med_ner.living_species.roberta
Code:
nlu.load('es.med_ner.living_species.roberta').predict('Lactante varón de dos años. Antecedentes familiares sin interés. Antecedentes personales: Embarazo, parto y periodo neonatal normal. En seguimiento por alergia a legumbres, diagnosticado con diez meses por reacción urticarial generalizada con lentejas y garbanzos, con dieta de exclusión a legumbres desde entonces. En ésta visita la madre describe episodios de eritema en zona maxilar derecha con afectación ocular ipsilateral que se resuelve en horas tras la administración de corticoides. Le ha ocurrido en 5-6 ocasiones, en relación con la ingesta de alimentos previamente tolerados. Exploración complementaria: Cacahuete, ac(ige)19.2 Ku.arb/l. Resultados: Ante la sospecha clínica de Síndrome de Frey, se tranquiliza a los padres, explicándoles la naturaleza del cuadro y se cita para revisión anual.')
Results:
| entities_living_species | entities_living_species_class | entities_living_species_confidence | |
|---|---|---|---|
| 0 | Lactante varón | HUMAN | 0.93175 |
| 0 | familiares | HUMAN | 1 |
| 0 | personales | HUMAN | 1 |
| 0 | neonatal | HUMAN | 0.9997 |
| 0 | legumbres | SPECIES | 0.9962 |
| 0 | lentejas | SPECIES | 0.9988 |
| 0 | garbanzos | SPECIES | 0.9901 |
| 0 | legumbres | SPECIES | 0.9976 |
| 0 | madre | HUMAN | 1 |
| 0 | Cacahuete | SPECIES | 0.998 |
| 0 | padres | HUMAN | 1 |
Code:
nlu.load('es.med_ner.living_species.roberta').viz('Lactante varón de dos años. Antecedentes familiares sin interés. Antecedentes personales: Embarazo, parto y periodo neonatal normal. En seguimiento por alergia a legumbres, diagnosticado con diez meses por reacción urticarial generalizada con lentejas y garbanzos, con dieta de exclusión a legumbres desde entonces. En ésta visita la madre describe episodios de eritema en zona maxilar derecha con afectación ocular ipsilateral que se resuelve en horas tras la administración de corticoides. Le ha ocurrido en 5-6 ocasiones, en relación con la ingesta de alimentos previamente tolerados. Exploración complementaria: Cacahuete, ac(ige)19.2 Ku.arb/l. Resultados: Ante la sospecha clínica de Síndrome de Frey, se tranquiliza a los padres, explicándoles la naturaleza del cuadro y se cita para revisión anual.')
Results:

All healthcare models added in NLU 4.0 :
All NLU 4.0 Core Models
All core models added in NLU 4.0 : Can be found on the NLU website because of Github Limitations
Minor Improvements
- IOB Schema Detection for Tokenclassifiers and adding NER Converting in those cases
- Tweaks in column name generation of most annotators
Bug Fixes
- fixed bug in multi lang parsing
- fixed bug for Normalizers
- fixed bug in fetching metadata for resolvers
- fixed bug in deducting outputlevel and inferring output columns
- fixed broken nlp_refs
NLU Version 3.4.4
600 new models with over 75 new languages including Ancient,Dead and Extinct languages, 155 languages total covered, 400% Tokenizer Speedup, 18x USE-Embeddings GPU speedup in John Snow Labs NLU 3.4.4
We are very excited to announce NLU 3.4.4 has been released with over 600 new model, over 75 new languages and 155 languages covered in total,
400% speedup for tokenizers and 18x speedup of UniversalSentenceEncoder on GPU.
On the general NLP side we have transformer based Embeddings and Token Classifiers powered by state of the art CamemBertEmbeddings and DeBertaForTokenClassification based
architectures as well as various new models for
Historical, Ancient,Dead, Extinct, Genetic and Constructed languages like
Old Church Slavonic, Latin, Sanskrit, Esperanto, Volapük, Coptic, Nahuatl, Ancient Greek (to 1453), Old Russian.
On the healthcare side we have Portuguese De-identification Models, have NER models for Gene detection and finally RxNorm Sentence resolution model for mapping and extracting pharmaceutical actions (e.g. analgesic, hypoglycemic)
as well as treatments (e.g. backache, diabetes).
General NLP Models
All general NLP models
First time language models covered
The languages for these models are covered for the very first time ever by NLU.
| Number | Language Name(s) | NLU Reference | Spark NLP Reference | Task | Annotator Class | ISO-639-1 | ISO-639-2/639-5 | ISO-639-3 | Scope | Language Type |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Sanskrit | sa.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | sa | san | san | Individual | Ancient |
| 1 | Sanskrit | sa.lemma | lemma_vedic | Lemmatization | LemmatizerModel | sa | san | san | Individual | Ancient |
| 2 | Sanskrit | sa.pos | pos_vedic | Part of Speech Tagging | PerceptronModel | sa | san | san | Individual | Ancient |
| 3 | Sanskrit | sa.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | sa | san | san | Individual | Ancient |
| 4 | Volapük | vo.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | vo | vol | vol | Individual | Constructed |
| 5 | Nahuatl languages | nah.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nah | nan | Collective | Genetic |
| 6 | Aragonese | an.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | an | arg | arg | Individual | Living |
| 7 | Assamese | as.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | as | asm | asm | Individual | Living |
| 8 | Asturian, Asturleonese, Bable, Leonese | ast.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | ast | ast | Individual | Living |
| 9 | Bashkir | ba.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | ba | bak | bak | Individual | Living |
| 10 | Bavarian | bar.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | bar | Individual | Living |
| 11 | Bishnupriya | bpy.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | bpy | Individual | Living |
| 12 | Burmese | my.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | my | 639-2/T: mya639-2/B: bur | mya | Individual | Living |
| 13 | Cebuano | ceb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | ceb | ceb | Individual | Living |
| 14 | Central Bikol | bcl.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | bcl | Individual | Living |
| 15 | Chechen | ce.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | ce | che | che | Individual | Living |
| 16 | Chuvash | cv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | cv | chv | chv | Individual | Living |
| 17 | Corsican | co.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | co | cos | cos | Individual | Living |
| 18 | Dhivehi, Divehi, Maldivian | dv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | dv | div | div | Individual | Living |
| 19 | Egyptian Arabic | arz.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | arz | Individual | Living |
| 20 | Emiliano-Romagnolo | eml.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | eml | nan | nan | Individual | Living |
| 21 | Erzya | myv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | myv | myv | Individual | Living |
| 22 | Georgian | ka.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | ka | 639-2/T: kat639-2/B: geo | kat | Individual | Living |
| 23 | Goan Konkani | gom.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | gom | Individual | Living |
| 24 | Javanese | jv.embed.distilbert | distilbert_embeddings_javanese_distilbert_small | Embeddings | DistilBertEmbeddings | jv | jav | jav | Individual | Living |
| 25 | Javanese | jv.embed.javanese_distilbert_small_imdb | distilbert_embeddings_javanese_distilbert_small_imdb | Embeddings | DistilBertEmbeddings | jv | jav | jav | Individual | Living |
| 26 | Javanese | jv.embed.javanese_roberta_small | roberta_embeddings_javanese_roberta_small | Embeddings | RoBertaEmbeddings | jv | jav | jav | Individual | Living |
| 27 | Javanese | jv.embed.javanese_roberta_small_imdb | roberta_embeddings_javanese_roberta_small_imdb | Embeddings | RoBertaEmbeddings | jv | jav | jav | Individual | Living |
| 28 | Javanese | jv.embed.javanese_bert_small_imdb | bert_embeddings_javanese_bert_small_imdb | Embeddings | BertEmbeddings | jv | jav | jav | Individual | Living |
| 29 | Javanese | jv.embed.javanese_bert_small | bert_embeddings_javanese_bert_small | Embeddings | BertEmbeddings | jv | jav | jav | Individual | Living |
| 30 | Kirghiz, Kyrgyz | ky.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | ky | kir | kir | Individual | Living |
| 31 | Letzeburgesch, Luxembourgish | lb.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | lb | ltz | ltz | Individual | Living |
| 32 | Letzeburgesch, Luxembourgish | lb.lemma | lemma_spacylookup | Lemmatization | LemmatizerModel | lb | ltz | ltz | Individual | Living |
| 33 | Letzeburgesch, Luxembourgish | lb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | lb | ltz | ltz | Individual | Living |
| 34 | Ligurian | lij.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | nan | nan | lij | Individual | Living |
| 35 | Lombard | lmo.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | lmo | Individual | Living |
| 36 | Low German, Low Saxon | nds.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nds | nds | Individual | Living |
| 37 | Macedonian | mk.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | mk | 639-2/T: mkd639-2/B: mac | mkd | Individual | Living |
| 38 | Macedonian | mk.lemma | lemma_spacylookup | Lemmatization | LemmatizerModel | mk | 639-2/T: mkd639-2/B: mac | mkd | Individual | Living |
| 39 | Macedonian | mk.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | mk | 639-2/T: mkd639-2/B: mac | mkd | Individual | Living |
| 40 | Maithili | mai.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | mai | mai | Individual | Living |
| 41 | Manx | gv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | gv | glv | glv | Individual | Living |
| 42 | Mazanderani | mzn.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | mzn | Individual | Living |
| 43 | Minangkabau | min.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | min | min | Individual | Living |
| 44 | Mingrelian | xmf.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | xmf | Individual | Living |
| 45 | Mirandese | mwl.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | mwl | mwl | Individual | Living |
| 46 | Neapolitan | nap.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nap | nap | Individual | Living |
| 47 | Nepal Bhasa, Newari | new.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | new | new | Individual | Living |
| 48 | Northern Frisian | frr.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | frr | frr | Individual | Living |
| 49 | Northern Sami | sme.lemma | lemma_giella | Lemmatization | LemmatizerModel | se | sme | sme | Individual | Living |
| 50 | Northern Sami | sme.pos | pos_giella | Part of Speech Tagging | PerceptronModel | se | sme | sme | Individual | Living |
| 51 | Northern Sotho, Pedi, Sepedi | nso.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nso | nso | Individual | Living |
| 52 | Occitan (post 1500) | oc.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | oc | oci | oci | Individual | Living |
| 53 | Ossetian, Ossetic | os.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | os | oss | oss | Individual | Living |
| 54 | Pfaelzisch | pfl.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | pfl | Individual | Living |
| 55 | Piemontese | pms.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | pms | Individual | Living |
| 56 | Romansh | rm.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | rm | roh | roh | Individual | Living |
| 57 | Scots | sco.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | sco | sco | Individual | Living |
| 58 | Sicilian | scn.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | scn | scn | Individual | Living |
| 59 | Sinhala, Sinhalese | si.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | si | sin | sin | Individual | Living |
| 60 | Sinhala, Sinhalese | si.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | si | sin | sin | Individual | Living |
| 61 | Sundanese | su.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | su | sun | sun | Individual | Living |
| 62 | Sundanese | su.embed.sundanese_roberta_base | roberta_embeddings_sundanese_roberta_base | Embeddings | RoBertaEmbeddings | su | sun | sun | Individual | Living |
| 63 | Tagalog | tl.lemma | lemma_spacylookup | Lemmatization | LemmatizerModel | tl | tgl | tgl | Individual | Living |
| 64 | Tagalog | tl.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | tl | tgl | tgl | Individual | Living |
| 65 | Tagalog | tl.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | tl | tgl | tgl | Individual | Living |
| 66 | Tagalog | tl.embed.roberta_tagalog_large | roberta_embeddings_roberta_tagalog_large | Embeddings | RoBertaEmbeddings | tl | tgl | tgl | Individual | Living |
| 67 | Tagalog | tl.embed.roberta_tagalog_base | roberta_embeddings_roberta_tagalog_base | Embeddings | RoBertaEmbeddings | tl | tgl | tgl | Individual | Living |
| 68 | Tajik | tg.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | tg | tgk | tgk | Individual | Living |
| 69 | Tatar | tt.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | tt | tat | tat | Individual | Living |
| 70 | Tatar | tt.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | tt | tat | tat | Individual | Living |
| 71 | Tigrinya | ti.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | ti | tir | tir | Individual | Living |
| 72 | Tosk Albanian | als.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | als | Individual | Living |
| 73 | Tswana | tn.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | tn | tsn | tsn | Individual | Living |
| 74 | Turkmen | tk.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | tk | tuk | tuk | Individual | Living |
| 75 | Upper Sorbian | hsb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | hsb | hsb | Individual | Living |
| 76 | Venetian | vec.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | vec | Individual | Living |
| 77 | Vlaams | vls.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | vls | Individual | Living |
| 78 | Walloon | wa.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | wa | wln | wln | Individual | Living |
| 79 | Waray (Philippines) | war.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | war | war | Individual | Living |
| 80 | Western Armenian | hyw.pos | pos_armtdp | Part of Speech Tagging | PerceptronModel | nan | nan | hyw | Individual | Living |
| 81 | Western Armenian | hyw.lemma | lemma_armtdp | Lemmatization | LemmatizerModel | nan | nan | hyw | Individual | Living |
| 82 | Western Frisian | fy.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | fy | fry | fry | Individual | Living |
| 83 | Western Panjabi | pnb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | pnb | Individual | Living |
| 84 | Yakut | sah.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | sah | sah | Individual | Living |
| 85 | Zeeuws | zea.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | nan | nan | zea | Individual | Living |
| 86 | Albanian | sq.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | sq | 639-2/T: sqi639-2/B: alb | sqi | Macrolanguage | Living |
| 87 | Albanian | sq.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | sq | 639-2/T: sqi639-2/B: alb | sqi | Macrolanguage | Living |
| 88 | Azerbaijani | az.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | az | aze | aze | Macrolanguage | Living |
| 89 | Azerbaijani | az.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | az | aze | aze | Macrolanguage | Living |
| 90 | Malagasy | mg.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | mg | mlg | mlg | Macrolanguage | Living |
| 91 | Malay (macrolanguage) | ms.embed.albert | albert_embeddings_albert_large_bahasa_cased | Embeddings | AlbertEmbeddings | ms | 639-2/T: msa639-2/B: may | msa | Macrolanguage | Living |
| 92 | Malay (macrolanguage) | ms.embed.distilbert | distilbert_embeddings_malaysian_distilbert_small | Embeddings | DistilBertEmbeddings | ms | 639-2/T: msa639-2/B: may | msa | Macrolanguage | Living |
| 93 | Malay (macrolanguage) | ms.embed.albert_tiny_bahasa_cased | albert_embeddings_albert_tiny_bahasa_cased | Embeddings | AlbertEmbeddings | ms | 639-2/T: msa639-2/B: may | msa | Macrolanguage | Living |
| 94 | Malay (macrolanguage) | ms.embed.albert_base_bahasa_cased | albert_embeddings_albert_base_bahasa_cased | Embeddings | AlbertEmbeddings | ms | 639-2/T: msa639-2/B: may | msa | Macrolanguage | Living |
| 95 | Malay (macrolanguage) | ms.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | ms | 639-2/T: msa639-2/B: may | msa | Macrolanguage | Living |
| 96 | Mongolian | mn.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | mn | mon | mon | Macrolanguage | Living |
| 97 | Oriya (macrolanguage) | or.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | or | ori | ori | Macrolanguage | Living |
| 98 | Pashto, Pushto | ps.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | ps | pus | pus | Macrolanguage | Living |
| 99 | Quechua | qu.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | qu | que | que | Macrolanguage | Living |
| 100 | Sardinian | sc.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | sc | srd | srd | Macrolanguage | Living |
| 101 | Serbo-Croatian | sh.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | sh | nan | nan | Macrolanguage | Living |
| 102 | Uzbek | uz.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | uz | uzb | uzb | Macrolanguage | Living |
All general NLP models
Powered by the incredible Spark NLP 3.4.4 and previous releases.
All Healthcare
Powered by the amazing Spark NLP for Healthcare 3.5.2 and Spark NLP for Healthcare 3.5.1 releases.
| Number | NLU Reference | Spark NLP Reference | Task | Language Name(s) | Annotator Class | ISO-639-1 | ISO-639-2/639-5 | ISO-639-3 | Language Type | Scope |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | en.med_ner.biomedical_bc2gm | ner_biomedical_bc2gm | Named Entity Recognition | English | MedicalNerModel | en | eng | eng | Living | Individual |
| 1 | en.med_ner.biomedical_bc2gm | ner_biomedical_bc2gm | Named Entity Recognition | English | MedicalNerModel | en | eng | eng | Living | Individual |
| 2 | en.resolve.rxnorm_action_treatment | sbiobertresolve_rxnorm_action_treatment | Entity Resolution | English | SentenceEntityResolverModel | en | eng | eng | Living | Individual |
| 3 | en.classify.token_bert.ner_ade | bert_token_classifier_ner_ade | Named Entity Recognition | English | MedicalBertForTokenClassifier | en | eng | eng | Living | Individual |
| 4 | en.classify.token_bert.ner_ade | bert_token_classifier_ner_ade | Named Entity Recognition | English | MedicalBertForTokenClassifier | en | eng | eng | Living | Individual |
| 5 | pt.med_ner.deid.subentity | ner_deid_subentity | De-identification | Portuguese | MedicalNerModel | pt | por | por | Living | Individual |
| 6 | pt.med_ner.deid.generic | ner_deid_generic | De-identification | Portuguese | MedicalNerModel | pt | por | por | Living | Individual |
| 7 | pt.med_ner.deid | ner_deid_generic | De-identification | Portuguese | MedicalNerModel | pt | por | por | Living | Individual |
NLU Version 3.4.3
Zero-Shot-Relation-Extraction, DeBERTa for Sequence Classification, 150+ new models, 60+ Languages in John Snow Labs NLU 3.4.3
We are very excited to announce NLU 3.4.3 has been released!
This release features new models for Zero-Shot-Relation-Extraction, DeBERTa for Sequence Classification,
Deidentification in French and Italian and
Lemmatizers, Parts of Speech Taggers, and Word2Vec Embeddings for over 66 languages, with 20 languages being covered
for the first time by NLU, including ancient and exotic languages like Ancient Greek, Old Russian,
Old French and much more. Once again we would like to thank our community to make this release possible.
NLU for Healthcare
On the healthcare NLP side, a new ZeroShotRelationExtractionModel is available, which can extract relations between
clinical entities in an unsupervised fashion, no training required!
Additionally, New French and Italian Deidentification models are available for clinical and healthcare domains.
Powerd by the fantastic Spark NLP for healthcare 3.5.0 release
Zero-Shot Relation Extraction
Zero-shot Relation Extraction to extract relations between clinical entities with no training dataset
import nlu
pipe = nlu.load('med_ner.clinical relation.zeroshot_biobert')
# Configure relations to extract
pipe['zero_shot_relation_extraction'].setRelationalCategories({
"CURE": [" cures ."],
"IMPROVE": [" improves .", " cures ."],
"REVEAL": [" reveals ."]})
.setMultiLabel(False)
df = pipe.predict("Paracetamol can alleviate headache or sickness. An MRI test can be used to find cancer.")
df[
'relation', 'relation_confidence', 'relation_entity1', 'relation_entity1_class', 'relation_entity2', 'relation_entity2_class',]
# Results in following table :
| relation | relation_confidence | relation_entity1 | relation_entity1_class | relation_entity2 | relation_entity2_class |
|---|---|---|---|---|---|
| REVEAL | 0.976004 | An MRI test | TEST | cancer | PROBLEM |
| IMPROVE | 0.988195 | Paracetamol | TREATMENT | sickness | PROBLEM |
| IMPROVE | 0.992962 | Paracetamol | TREATMENT | headache | PROBLEM |
New Healthcare Models overview
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.relation.zeroshot_biobert | re_zeroshot_biobert | Relation Extraction | ZeroShotRelationExtractionModel |
| fr | fr.med_ner.deid_generic | ner_deid_generic | De-identification | MedicalNerModel |
| fr | fr.med_ner.deid_subentity | ner_deid_subentity | De-identification | MedicalNerModel |
| it | it.med_ner.deid_generic | ner_deid_generic | Named Entity Recognition | MedicalNerModel |
| it | it.med_ner.deid_subentity | ner_deid_subentity | Named Entity Recognition | MedicalNerModel |
NLU general
On the general NLP side we have new transformer based DeBERTa v3 sequence classifiers models fine-tuned in Urdu, French and English for
Sentiment and News classification. Additionally, 100+ Part Of Speech Taggers and Lemmatizers for 66 Languages and for 7
languages new word2vec embeddings, including hi,azb,bo,diq,cy,es,it,
powered by the amazing Spark NLP 3.4.3 release
New Languages covered:
First time languages covered by NLU are :
South Azerbaijani, Tibetan, Dimli, Central Kurdish, Southern Altai,
Scottish Gaelic,Faroese,Literary Chinese,Ancient Greek,
Gothic, Old Russian, Church Slavic,
Old French,Uighur,Coptic,Croatian, Belarusian, Serbian
and their respective ISO-639-3 and ISO 630-2 codes are :
azb,bo,diq,ckb, lt gd, fo,lzh,grc,got,orv,cu,fro,qtd,ug,cop,hr,be,qhe,sr
New NLP Models Overview
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.classify.sentiment.imdb.deberta | deberta_v3_xsmall_sequence_classifier_imdb | Text Classification | DeBertaForSequenceClassification |
| en | en.classify.sentiment.imdb.deberta.small | deberta_v3_small_sequence_classifier_imdb | Text Classification | DeBertaForSequenceClassification |
| en | en.classify.sentiment.imdb.deberta.base | deberta_v3_base_sequence_classifier_imdb | Text Classification | DeBertaForSequenceClassification |
| en | en.classify.sentiment.imdb.deberta.large | deberta_v3_large_sequence_classifier_imdb | Text Classification | DeBertaForSequenceClassification |
| en | en.classify.news.deberta | deberta_v3_xsmall_sequence_classifier_ag_news | Text Classification | DeBertaForSequenceClassification |
| en | en.classify.news.deberta.small | deberta_v3_small_sequence_classifier_ag_news | Text Classification | DeBertaForSequenceClassification |
| ur | ur.classify.sentiment.imdb | mdeberta_v3_base_sequence_classifier_imdb | Text Classification | DeBertaForSequenceClassification |
| fr | fr.classify.allocine | mdeberta_v3_base_sequence_classifier_allocine | Text Classification | DeBertaForSequenceClassification |
| ur | ur.embed.bert_cased | bert_embeddings_bert_base_ur_cased | Embeddings | BertEmbeddings |
| fr | fr.embed.bert_5lang_cased | bert_embeddings_bert_base_5lang_cased | Embeddings | BertEmbeddings |
| de | de.embed.medbert | bert_embeddings_German_MedBERT | Embeddings | BertEmbeddings |
| ar | ar.embed.arbert | bert_embeddings_ARBERT | Embeddings | BertEmbeddings |
| bn | bn.embed.bangala_bert | bert_embeddings_bangla_bert_base | Embeddings | BertEmbeddings |
| zh | zh.embed.bert_5lang_cased | bert_embeddings_bert_base_5lang_cased | Embeddings | BertEmbeddings |
| hi | hi.embed.bert_hi_cased | bert_embeddings_bert_base_hi_cased | Embeddings | BertEmbeddings |
| it | it.embed.bert_it_cased | bert_embeddings_bert_base_it_cased | Embeddings | BertEmbeddings |
| ko | ko.embed.bert | bert_embeddings_bert_base | Embeddings | BertEmbeddings |
| tr | tr.embed.bert_cased | bert_embeddings_bert_base_tr_cased | Embeddings | BertEmbeddings |
| vi | vi.embed.bert_cased | bert_embeddings_bert_base_vi_cased | Embeddings | BertEmbeddings |
| hif | hif.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| azb | azb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| bo | bo.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| diq | diq.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| cy | cy.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| es | es.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| it | it.embed.word2vec | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| af | af.lemma | lemma | Lemmatization | LemmatizerModel |
| lt | lt.lemma | lemma_alksnis | Lemmatization | LemmatizerModel |
| nl | nl.lemma | lemma | Lemmatization | LemmatizerModel |
| gd | gd.lemma | lemma_arcosg | Lemmatization | LemmatizerModel |
| es | es.lemma | lemma | Lemmatization | LemmatizerModel |
| ca | ca.lemma | lemma | Lemmatization | LemmatizerModel |
| el | el.lemma.gdt | lemma_gdt | Lemmatization | LemmatizerModel |
| en | en.lemma.atis | lemma_atis | Lemmatization | LemmatizerModel |
| tr | tr.lemma.boun | lemma_boun | Lemmatization | LemmatizerModel |
| da | da.lemma.ddt | lemma_ddt | Lemmatization | LemmatizerModel |
| cs | cs.lemma.cac | lemma_cac | Lemmatization | LemmatizerModel |
| en | en.lemma.esl | lemma_esl | Lemmatization | LemmatizerModel |
| bg | bg.lemma.btb | lemma_btb | Lemmatization | LemmatizerModel |
| id | id.lemma.csui | lemma_csui | Lemmatization | LemmatizerModel |
| gl | gl.lemma.ctg | lemma_ctg | Lemmatization | LemmatizerModel |
| cy | cy.lemma.ccg | lemma_ccg | Lemmatization | LemmatizerModel |
| fo | fo.lemma.farpahc | lemma_farpahc | Lemmatization | LemmatizerModel |
| tr | tr.lemma.atis | lemma_atis | Lemmatization | LemmatizerModel |
| ga | ga.lemma.idt | lemma_idt | Lemmatization | LemmatizerModel |
| ja | ja.lemma.gsdluw | lemma_gsdluw | Lemmatization | LemmatizerModel |
| es | es.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| en | en.lemma.gum | lemma_gum | Lemmatization | LemmatizerModel |
| zh | zh.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| lv | lv.lemma.lvtb | lemma_lvtb | Lemmatization | LemmatizerModel |
| hi | hi.lemma.hdtb | lemma_hdtb | Lemmatization | LemmatizerModel |
| pt | pt.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| de | de.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| nl | nl.lemma.lassysmall | lemma_lassysmall | Lemmatization | LemmatizerModel |
| lzh | lzh.lemma.kyoto | lemma_kyoto | Lemmatization | LemmatizerModel |
| zh | zh.lemma.gsdsimp | lemma_gsdsimp | Lemmatization | LemmatizerModel |
| he | he.lemma.htb | lemma_htb | Lemmatization | LemmatizerModel |
| fr | fr.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| ro | ro.lemma.nonstandard | lemma_nonstandard | Lemmatization | LemmatizerModel |
| ja | ja.lemma.gsd | lemma_gsd | Lemmatization | LemmatizerModel |
| it | it.lemma.isdt | lemma_isdt | Lemmatization | LemmatizerModel |
| de | de.lemma.hdt | lemma_hdt | Lemmatization | LemmatizerModel |
| is | is.lemma.modern | lemma_modern | Lemmatization | LemmatizerModel |
| la | la.lemma.ittb | lemma_ittb | Lemmatization | LemmatizerModel |
| fr | fr.lemma.partut | lemma_partut | Lemmatization | LemmatizerModel |
| pcm | pcm.lemma.nsc | lemma_nsc | Lemmatization | LemmatizerModel |
| pl | pl.lemma.pdb | lemma_pdb | Lemmatization | LemmatizerModel |
| grc | grc.lemma.perseus | lemma_perseus | Lemmatization | LemmatizerModel |
| cs | cs.lemma.pdt | lemma_pdt | Lemmatization | LemmatizerModel |
| fa | fa.lemma.perdt | lemma_perdt | Lemmatization | LemmatizerModel |
| got | got.lemma.proiel | lemma_proiel | Lemmatization | LemmatizerModel |
| fr | fr.lemma.rhapsodie | lemma_rhapsodie | Lemmatization | LemmatizerModel |
| it | it.lemma.partut | lemma_partut | Lemmatization | LemmatizerModel |
| en | en.lemma.partut | lemma_partut | Lemmatization | LemmatizerModel |
| no | no.lemma.nynorsklia | lemma_nynorsklia | Lemmatization | LemmatizerModel |
| orv | orv.lemma.rnc | lemma_rnc | Lemmatization | LemmatizerModel |
| cu | cu.lemma.proiel | lemma_proiel | Lemmatization | LemmatizerModel |
| la | la.lemma.perseus | lemma_perseus | Lemmatization | LemmatizerModel |
| fr | fr.lemma.parisstories | lemma_parisstories | Lemmatization | LemmatizerModel |
| fro | fro.lemma.srcmf | lemma_srcmf | Lemmatization | LemmatizerModel |
| vi | vi.lemma.vtb | lemma_vtb | Lemmatization | LemmatizerModel |
| qtd | qtd.lemma.sagt | lemma_sagt | Lemmatization | LemmatizerModel |
| ro | ro.lemma.rrt | lemma_rrt | Lemmatization | LemmatizerModel |
| hu | hu.lemma.szeged | lemma_szeged | Lemmatization | LemmatizerModel |
| ug | ug.lemma.udt | lemma_udt | Lemmatization | LemmatizerModel |
| wo | wo.lemma.wtb | lemma_wtb | Lemmatization | LemmatizerModel |
| cop | cop.lemma.scriptorium | lemma_scriptorium | Lemmatization | LemmatizerModel |
| ru | ru.lemma.syntagrus | lemma_syntagrus | Lemmatization | LemmatizerModel |
| ru | ru.lemma.taiga | lemma_taiga | Lemmatization | LemmatizerModel |
| fr | fr.lemma.sequoia | lemma_sequoia | Lemmatization | LemmatizerModel |
| la | la.lemma.udante | lemma_udante | Lemmatization | LemmatizerModel |
| ro | ro.lemma.simonero | lemma_simonero | Lemmatization | LemmatizerModel |
| it | it.lemma.vit | lemma_vit | Lemmatization | LemmatizerModel |
| hr | hr.lemma.set | lemma_set | Lemmatization | LemmatizerModel |
| fa | fa.lemma.seraji | lemma_seraji | Lemmatization | LemmatizerModel |
| tr | tr.lemma.tourism | lemma_tourism | Lemmatization | LemmatizerModel |
| ta | ta.lemma.ttb | lemma_ttb | Lemmatization | LemmatizerModel |
| sl | sl.lemma.ssj | lemma_ssj | Lemmatization | LemmatizerModel |
| sv | sv.lemma.talbanken | lemma_talbanken | Lemmatization | LemmatizerModel |
| uk | uk.lemma.iu | lemma_iu | Lemmatization | LemmatizerModel |
| te | te.pos | pos_mtg | Part of Speech Tagging | PerceptronModel |
| te | te.pos | pos_mtg | Part of Speech Tagging | PerceptronModel |
| ta | ta.pos | pos_ttb | Part of Speech Tagging | PerceptronModel |
| ta | ta.pos | pos_ttb | Part of Speech Tagging | PerceptronModel |
| cs | cs.pos | pos_ud_pdt | Part of Speech Tagging | PerceptronModel |
| cs | cs.pos | pos_ud_pdt | Part of Speech Tagging | PerceptronModel |
| bg | bg.pos | pos_btb | Part of Speech Tagging | PerceptronModel |
| bg | bg.pos | pos_btb | Part of Speech Tagging | PerceptronModel |
| af | af.pos | pos_afribooms | Part of Speech Tagging | PerceptronModel |
| af | af.pos | pos_afribooms | Part of Speech Tagging | PerceptronModel |
| af | af.pos | pos_afribooms | Part of Speech Tagging | PerceptronModel |
| es | es.pos.gsd | pos_gsd | Part of Speech Tagging | PerceptronModel |
| en | en.pos.ewt | pos_ewt | Part of Speech Tagging | PerceptronModel |
| gd | gd.pos.arcosg | pos_arcosg | Part of Speech Tagging | PerceptronModel |
| el | el.pos.gdt | pos_gdt | Part of Speech Tagging | PerceptronModel |
| hy | hy.pos.armtdp | pos_armtdp | Part of Speech Tagging | PerceptronModel |
| pt | pt.pos.bosque | pos_bosque | Part of Speech Tagging | PerceptronModel |
| tr | tr.pos.framenet | pos_framenet | Part of Speech Tagging | PerceptronModel |
| cs | cs.pos.cltt | pos_cltt | Part of Speech Tagging | PerceptronModel |
| eu | eu.pos.bdt | pos_bdt | Part of Speech Tagging | PerceptronModel |
| et | et.pos.ewt | pos_ewt | Part of Speech Tagging | PerceptronModel |
| da | da.pos.ddt | pos_ddt | Part of Speech Tagging | PerceptronModel |
| cy | cy.pos.ccg | pos_ccg | Part of Speech Tagging | PerceptronModel |
| lt | lt.pos.alksnis | pos_alksnis | Part of Speech Tagging | PerceptronModel |
| nl | nl.pos.alpino | pos_alpino | Part of Speech Tagging | PerceptronModel |
| fi | fi.pos.ftb | pos_ftb | Part of Speech Tagging | PerceptronModel |
| tr | tr.pos.atis | pos_atis | Part of Speech Tagging | PerceptronModel |
| ca | ca.pos.ancora | pos_ancora | Part of Speech Tagging | PerceptronModel |
| gl | gl.pos.ctg | pos_ctg | Part of Speech Tagging | PerceptronModel |
| de | de.pos.gsd | pos_gsd | Part of Speech Tagging | PerceptronModel |
| fr | fr.pos.gsd | pos_gsd | Part of Speech Tagging | PerceptronModel |
| ja | ja.pos.gsdluw | pos_gsdluw | Part of Speech Tagging | PerceptronModel |
| it | it.pos.isdt | pos_isdt | Part of Speech Tagging | PerceptronModel |
| be | be.pos.hse | pos_hse | Part of Speech Tagging | PerceptronModel |
| nl | nl.pos.lassysmall | pos_lassysmall | Part of Speech Tagging | PerceptronModel |
| sv | sv.pos.lines | pos_lines | Part of Speech Tagging | PerceptronModel |
| uk | uk.pos.iu | pos_iu | Part of Speech Tagging | PerceptronModel |
| fr | fr.pos.parisstories | pos_parisstories | Part of Speech Tagging | PerceptronModel |
| en | en.pos.partut | pos_partut | Part of Speech Tagging | PerceptronModel |
| la | la.pos.ittb | pos_ittb | Part of Speech Tagging | PerceptronModel |
| lzh | lzh.pos.kyoto | pos_kyoto | Part of Speech Tagging | PerceptronModel |
| id | id.pos.gsd | pos_gsd | Part of Speech Tagging | PerceptronModel |
| he | he.pos.htb | pos_htb | Part of Speech Tagging | PerceptronModel |
| tr | tr.pos.kenet | pos_kenet | Part of Speech Tagging | PerceptronModel |
| de | de.pos.hdt | pos_hdt | Part of Speech Tagging | PerceptronModel |
| qhe | qhe.pos.hiencs | pos_hiencs | Part of Speech Tagging | PerceptronModel |
| la | la.pos.llct | pos_llct | Part of Speech Tagging | PerceptronModel |
| en | en.pos.lines | pos_lines | Part of Speech Tagging | PerceptronModel |
| pcm | pcm.pos.nsc | pos_nsc | Part of Speech Tagging | PerceptronModel |
| ko | ko.pos.kaist | pos_kaist | Part of Speech Tagging | PerceptronModel |
| pt | pt.pos.gsd | pos_gsd | Part of Speech Tagging | PerceptronModel |
| hi | hi.pos.hdtb | pos_hdtb | Part of Speech Tagging | PerceptronModel |
| is | is.pos.modern | pos_modern | Part of Speech Tagging | PerceptronModel |
| en | en.pos.gum | pos_gum | Part of Speech Tagging | PerceptronModel |
| fro | fro.pos.srcmf | pos_srcmf | Part of Speech Tagging | PerceptronModel |
| sl | sl.pos.ssj | pos_ssj | Part of Speech Tagging | PerceptronModel |
| ru | ru.pos.taiga | pos_taiga | Part of Speech Tagging | PerceptronModel |
| grc | grc.pos.perseus | pos_perseus | Part of Speech Tagging | PerceptronModel |
| sr | sr.pos.set | pos_set | Part of Speech Tagging | PerceptronModel |
| orv | orv.pos.rnc | pos_rnc | Part of Speech Tagging | PerceptronModel |
| ug | ug.pos.udt | pos_udt | Part of Speech Tagging | PerceptronModel |
| got | got.pos.proiel | pos_proiel | Part of Speech Tagging | PerceptronModel |
| sv | sv.pos.talbanken | pos_talbanken | Part of Speech Tagging | PerceptronModel |
| sv | sv.pos.talbanken | pos_talbanken | Part of Speech Tagging | PerceptronModel |
| pl | pl.pos.pdb | pos_pdb | Part of Speech Tagging | PerceptronModel |
| fa | fa.pos.seraji | pos_seraji | Part of Speech Tagging | PerceptronModel |
| tr | tr.pos.penn | pos_penn | Part of Speech Tagging | PerceptronModel |
| hu | hu.pos.szeged | pos_szeged | Part of Speech Tagging | PerceptronModel |
| sk | sk.pos.snk | pos_snk | Part of Speech Tagging | PerceptronModel |
| sk | sk.pos.snk | pos_snk | Part of Speech Tagging | PerceptronModel |
| ro | ro.pos.simonero | pos_simonero | Part of Speech Tagging | PerceptronModel |
| it | it.pos.postwita | pos_postwita | Part of Speech Tagging | PerceptronModel |
| gl | gl.pos.treegal | pos_treegal | Part of Speech Tagging | PerceptronModel |
| cs | cs.pos.pdt | pos_pdt | Part of Speech Tagging | PerceptronModel |
| ro | ro.pos.rrt | pos_rrt | Part of Speech Tagging | PerceptronModel |
| orv | orv.pos.torot | pos_torot | Part of Speech Tagging | PerceptronModel |
| hr | hr.pos.set | pos_set | Part of Speech Tagging | PerceptronModel |
| la | la.pos.proiel | pos_proiel | Part of Speech Tagging | PerceptronModel |
| fr | fr.pos.partut | pos_partut | Part of Speech Tagging | PerceptronModel |
| it | it.pos.vit | pos_vit | Part of Speech Tagging | PerceptronModel |
Bugfixes
- Improved Error Messages and integrated detection and stopping of endless loops which could occur during construction of nlu pipelines
Additional NLU resources
- 140+ NLU Tutorials
- NLU in Action
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.4.2
Multilingual DeBERTa Transformer Embeddings for 100+ Languages, Spanish Deidentification and NER for Randomized Clinical Trials - John Snow Labs NLU 3.4.2
We are very excited NLU 3.4.2 has been released. On the open source side we have 5 new DeBERTa Transformer models for English and Multi-Lingual for 100+ languages. DeBERTa improves over BERT and RoBERTa by introducing two novel techniques.
For the healthcare side we have new NER models for randomized clinical trials (RCT) which can detect entities of type
BACKGROUND, CONCLUSIONS, METHODS, OBJECTIVE, RESULTS from clinical text.
Additionally, new Spanish Deidentification NER models for entities like STATE, PATIENT, DEVICE, COUNTRY, ZIP, PHONE, HOSPITAL and many more.
New Open Source Models
Integrates models from Spark NLP 3.4.2 release
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.embed.deberta_v3_xsmall | deberta_v3_xsmall | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_small | deberta_v3_small | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_base | deberta_v3_base | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_large | deberta_v3_large | Embeddings | DeBertaEmbeddings |
| xx | xx.embed.mdeberta_v3_base | mdeberta_v3_base | Embeddings | DeBertaEmbeddings |
New Healthcare Models
Integrates models from Spark NLP For Healthcare 3.4.2 release
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.med_ner.clinical_trials | bert_sequence_classifier_rct_biobert | Text Classification | MedicalBertForSequenceClassification |
| es | es.med_ner.deid.generic.roberta | ner_deid_generic_roberta_augmented | De-identification | MedicalNerModel |
| es | es.med_ner.deid.subentity.roberta | ner_deid_subentity_roberta_augmented | De-identification | MedicalNerModel |
| en | en.med_ner.deid.generic_augmented | ner_deid_generic_augmented | [‘Named Entity Recognition’, ‘De-identification’] | MedicalNerModel |
| en | en.med_ner.deid.subentity_augmented | ner_deid_subentity_augmented | [‘Named Entity Recognition’, ‘De-identification’] | MedicalNerModel |
Additional NLU resources
- 140+ NLU Tutorials
- NLU in Action
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.4.1
22 New models for 23 languages including various African and Indian languages, Medical Spanish models and more in NLU 3.4.1
We are very excited to announce the release of NLU 3.4.1 which features 22 new models for 23 languages where the The open-source side covers new Embeddings for Vietnamese and English Clinical domains and Multilingual Embeddings for 12 Indian and 9 African Languages. Additionally, there are new Sequence classifiers for Multilingual NER for 9 African languages, German Sentiment Classifiers and English Emotion and Typo Classifiers. The healthcare side covers Medical Spanish models, Classifiers for Drugs, Gender, the Pico Framework, and Relation Extractors for Adverse Drug events and Temporality. Finally, Spark 3.2.X is now supported and bugs related to Databricks environments have been fixed.
General NLU Improvements
- Support for Spark 3.2.x
New Open Source Models
Based on the amazing 3.4.1 Spark NLP Release integrates new Multilingual embeddings for 12 Major Indian languages, embeddings for Vietnamese, French, and English Clinical domains. Additionally new Multilingual NER model for 9 African languages, English 6 Class Emotion classifier and Typo detectors.
New Embeddings
- Multilingual ALBERT - IndicBert model pretrained exclusively on 12 major Indian languages with size smaller and performance on par or better than competing models. Languages covered are Assamese, Bengali, English, Gujarati, Hindi, Kannada, Malayalam, Marathi, Oriya, Punjabi, Tamil, Telugu. Available with xx.embed.albert.indic
- Fine tuned Vietnamese DistilBERT Base cased embeddings. Available with vi.embed.distilbert.cased
- Clinical Longformer Embeddings which consistently out-performs ClinicalBERT for various downstream tasks and on datasets. Available with en.embed.longformer.clinical
- Fine tuned Static French Word2Vec Embeddings in 3 sizes, 200d, 300d and 100d. Available with fr.embed.word2vec_wiki_1000, fr.embed.word2vec_wac_200 and fr.embed.w2v_cc_300d
New Transformer based Token and Sequence Classifiers
- Multilingual NER Distilbert model which detects entities
DATE,LOC,ORG,PERfor the languages 9 African languages (Hausa, Igbo, Kinyarwanda, Luganda, Nigerian, Pidgin, Swahili, Wolof, and Yorùbá). Available with xx.ner.masakhaner.distilbert - German News Sentiment Classifier available with de.classify.news_sentiment.bert
- English Emotion Classifier for 6 Classes available with en.classify.emotion.bert
- **English Typo Detector **: available with en.classify.typos.distilbert
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| xx | xx.embed.albert.indic | albert_indic | Embeddings | AlbertEmbeddings |
| xx | xx.ner.masakhaner.distilbert | xlm_roberta_large_token_classifier_masakhaner | Named Entity Recognition | DistilBertForTokenClassification |
| en | en.embed.longformer.clinical | clinical_longformer | Embeddings | LongformerEmbeddings |
| en | en.classify.emotion.bert | bert_sequence_classifier_emotion | Text Classification | BertForSequenceClassification |
| de | de.classify.news_sentiment.bert | bert_sequence_classifier_news_sentiment | Sentiment Analysis | BertForSequenceClassification |
| en | en.classify.typos.distilbert | distilbert_token_classifier_typo_detector | Named Entity Recognition | DistilBertForTokenClassification |
| fr | fr.embed.word2vec_wiki_1000 | word2vec_wiki_1000 | Embeddings | WordEmbeddingsModel |
| fr | fr.embed.word2vec_wac_200 | word2vec_wac_200 | Embeddings | WordEmbeddingsModel |
| fr | fr.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| vi | vi.embed.distilbert.cased | distilbert_base_cased | Embeddings | DistilBertEmbeddings |
New Healthcare Models
Integrated from the amazing 3.4.1 Spark NLP For Healthcare Release.
which makes 2 new Annotator Classes available, MedicalBertForSequenceClassification and MedicalDistilBertForSequenceClassification,
various medical Spanish models, RxNorm Resolvers,
Transformer based sequence classifiers for Drugs, Gender and the PICO framework,
and Relation extractors for Temporality and Causality of Drugs and Adverse Events.
New Medical Spanish Models
- Spanish Word2Vec Embeddings available with es.embed.sciwiki_300d
- Spanish PHI Deidentification NER models with two different subsets of entities extracted, available with ner_deid_generic and ner_deid_subentity
New Resolvers
- RxNorm resolvers with augmented concept data available with en.med_ner.supplement_clinical
New Transformer based Sequence Classifiers
- Adverse Drug Event Classifier Biobert based available with en.classify.ade.seq_biobert
- Patient Gender Classifier Biobert and Distilbert based available with en.classify.gender.seq_biobert and available with en.classify.ade.seq_distilbert
- PiCO Framework Classifier available with en.classify.pico.seq_biobert
New Relation Extractors
- Temporal Relation Extractor available with en.relation.temporal_events_clinical
- Adverse Drug Event Relation Extractors one version Biobert Embeddings and one non-DL version available with en.relation.adverse_drug_events.clinical available with en.relation.adverse_drug_events.clinical.biobert
Bugfixes
- Fixed bug that caused non-default output level of components to be sentence
- Fixed a bug that caused nlu references pointing to pretrained pipelines in spark nlp to crash in Databricks environments
Additional NLU resources
- 140+ NLU Tutorials
- NLU in Action
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.4.0
1 line to OCR for images, PDFS and DOCX, Text Generation with GPT2 and new T5 models, Sequence Classification with XlmRoBerta, RoBerta, Xlnet, Longformer and Albert, Transformer based medical NER with MedicalBertForTokenClassifier, 80 new models, 20+ new languages including various African and Scandinavian and much more in John Snow Labs NLU 3.4.0 !
We are incredibly excited to announce John Snow Labs NLU 3.4.0 has been released!
This release features 11 new annotator classes and 80 new models, including 3 OCR Transformers which enable you to extract text
from various file types, support for GPT2 and new pretrained T5 models for Text Generation and dozens more of new transformer based models
for Token and Sequence Classification.
This includes 8 new Sequence classifier models which can be pretrained in Huggingface and imported into Spark NLP and NLU.
Finally, the NLU tutorial page of the 140+ notebooks has been updated
New NLU OCR Features
3 new OCR based spells are supported, which enable extracting text from files of type
JPEG, PNG, BMP, WBMP, GIF, JPG, TIFF, DOCX, PDF in just 1 line of code.
You need a Spark OCR license for using these, which is available for free here and refer to the new
OCR tutorial notebook
Find more details on the NLU OCR documentation page
New NLU Healthcare Features
The healthcare side features a new MedicalBertForTokenClassifier annotator which is a Bert based model for token classification problems like Named Entity Recognition,
Parts of Speech and much more. Overall there are 28 new models which include German De-Identification models, English NER models for extracting Drug Development Trials,
Clinical Abbreviations and Acronyms, NER models for chemical compounds/drugs and genes/proteins, updated MedicalBertForTokenClassifier NER models for the medical domains Adverse drug Events,
Anatomy, Chemicals, Genes,Proteins, Cellular/Molecular Biology, Drugs, Bacteria, De-Identification and general Medical and Clinical Named Entities.
For Entity Relation Extraction between entity pairs new models for interaction between Drugs and Proteins.
For Entity Resolution new models for resolving Clinical Abbreviations and Acronyms to their full length names and also a model for resolving Drug Substance Entities to the categories
Clinical Drug, Pharmacologic Substance, Antibiotic, Hazardous or Poisonous Substance and new resolvers for LOINC and SNOMED terminologies.
New NLU Open source Features
On the open source side we have new support for Open Ai’s GPT2 for various text sequence to sequence problems and
additionally the following new Transformer models are supported :
RoBertaForSequenceClassification, XlmRoBertaForSequenceClassification, LongformerForSequenceClassification,
AlbertForSequenceClassification, XlnetForSequenceClassification, Word2Vec with various pre-trained weights for various problems!
New GPT2 models for generating text conditioned on some input,
New T5 style transfer models for active to passive, formal to informal, informal to formal, passive to active sequence to sequence generation.
Additionally, a new T5 model for generating SQL code from natural language input is provided.
On top of this dozens new Transformer based Sequence Classifiers and Token Classifiers have been released, this is includes for Token Classifier the following models :
Multi-Lingual general NER models for 10 African Languages (Amharic, Hausa, Igbo, Kinyarwanda, Luganda, Nigerian, Pidgin, Swahilu, Wolof, and Yorùbá),
10 high resourced languages (10 high resourced languages (Arabic, German, English, Spanish, French, Italian, Latvian, Dutch, Portuguese and Chinese),
6 Scandinavian languages (Danish, Norwegian-Bokmål, Norwegian-Nynorsk, Swedish, Icelandic, Faroese) ,
Uni-Lingual NER models for general entites in the language Chinese, Hindi, Islandic, Indonesian
and finally English NER models for extracting entities related to Stocks Ticker Symbols, Restaurants, Time.
For Sequence Classification new models for classifying Toxicity in Russian text and English models for
Movie Reviews, News Categorization, Sentimental Tone and General Sentiment
New NLU OCR Models
The following Transformers have been integrated from Spark OCR
| NLU Spell | Transformer Class |
|---|---|
nlu.load(img2text) |
ImageToText |
nlu.load(pdf2text) |
PdfToText |
nlu.load(doc2text) |
DocToText |
New Open Source Models
Integration for the 49 new models from the colossal Spark NLP 3.4.0 release
New Healthcare Models
Integration for the 28 new models from the amazing Spark NLP for healthcare 3.4.0 release
Additional NLU resources
- NLU OCR tutorial notebook
- 140+ NLU Tutorials
- NLU in Action
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.3.1
48 new Transformer based models in 9 new languages, including NER for Finance, Industry, Politcal Policies, COVID and Chemical Trials, various clinical and medical domains in Spanish and English and much more in NLU 3.3.1
We are incredibly excited to announce NLU 3.3.1 has been released with 48 new models in 9 languages!
It comes with 2 new types of state-of-the-art models,distilBERT and BERT for sequence classification with various pre-trained weights,
state-of-the-art bert based classifiers for problems in the domains of Finance, Sentiment Classification, Industry, News, and much more.
On the healthcare side, NLU features 22 new models in for English and Spanish with
with entity Resolver Models for LOINC, MeSH, NDC and SNOMED and UMLS Diseases,
NER models for Biomarkers, NIHSS-Guidelines, COVID Trials , Chemical Trials,
Bert based Token Classifier models for biological, genetical,cancer, cellular terms,
Bert for Sequence Classification models for clinical question vs statement classification
and finally Spanish Clinical NER and Resolver Models
Once again, we would like to thank our community for making another amazing release possible!
New Open Source Models and Features
Integrates the amazing Spark NLP 3.3.3 and 3.3.2 releases, featuring:
- New state-of-the-art fine-tuned
BERT models for Sequence ClassificationinEnglish,French,German,Spanish,Japanese,Turkish,Russian, and multilingual languages. DistilBertForSequenceClassificationmodels inEnglish,FrenchandUrduWord2Vecmodels.classify.distilbert_sequence.banking77:Banking NER modeltrained on BANKING77 dataset, which provides a very fine-grained set of intents in a banking domain. It comprises 13,083 customer service queries labeled with 77 intents. It focuses on fine-grained single-domain intent detection. Can extract entities like activate_my_card, age_limit, apple_pay_or_google_pay, atm_support, automatic_top_up, balance_not_updated_after_bank_transfer, balance_not_updated_after_cheque_or_cash_deposit, beneficiary_not_allowed, cancel_transfer, card_about_to_expire, card_acceptance, card_arrival, card_delivery_estimate, card_linking, card_not_working, card_payment_fee_charged, card_payment_not_recognised, card_payment_wrong_exchange_rate, card_swallowed, cash_withdrawal_charge, cash_withdrawal_not_recognised, change_pin, compromised_card, contactless_not_working, country_support, declined_card_payment, declined_cash_withdrawal, declined_transfer, direct_debit_payment_not_recognised, disposable_card_limits, edit_personal_details, exchange_charge, exchange_rate, exchange_via_app, extra_charge_on_statement, failed_transfer, fiat_currency_support, get_disposable_virtual_card, get_physical_card, getting_spare_card, getting_virtual_card, lost_or_stolen_card, lost_or_stolen_phone, order_physical_card, passcode_forgotten, pending_card_payment, pending_cash_withdrawal, pending_top_up, pending_transfer, pin_blocked, receiving_money,classify.distilbert_sequence.industry:Industry NER modelwhich can extract entities like Advertising, Aerospace & Defense, Apparel Retail, Apparel, Accessories & Luxury Goods, Application Software, Asset Management & Custody Banks, Auto Parts & Equipment, Biotechnology, Building Products, Casinos & Gaming, Commodity Chemicals, Communications Equipment, Construction & Engineering, Construction Machinery & Heavy Trucks, Consumer Finance, Data Processing & Outsourced Services, Diversified Metals & Mining, Diversified Support Services, Electric Utilities, Electrical Components & Equipment, Electronic Equipment & Instruments, Environmental & Facilities Services, Gold, Health Care Equipment, Health Care Facilities, Health Care Services.xx.classify.bert_sequence.sentiment:Multi-Lingual Sentiment ClassifierThis a bert-base-multilingual-uncased model finetuned for sentiment analysis on product reviews in six languages: English, Dutch, German, French, Spanish and Italian. It predicts the sentiment of the review as a number of stars (between 1 and 5). This model is intended for direct use as a sentiment analysis model for product reviews in any of the six languages above, or for further finetuning on related sentiment analysis tasks.distilbert_sequence.policy:Policy ClassifierThis model was trained on 129.669 manually annotated sentences to classify text into one of seven political categories: ‘Economy’, ‘External Relations’, ‘Fabric of Society’, ‘Freedom and Democracy’, ‘Political System’, ‘Welfare and Quality of Life’ or ‘Social Groups’.classify.bert_sequence.dehatebert_mono:Hate Speech ClassifierThis model was trained on 129.669 manually annotated sentences to classify text into one of seven political categories: ‘Economy’, ‘External Relations’, ‘Fabric of Society’, ‘Freedom and Democracy’, ‘Political System’, ‘Welfare and Quality of Life’ or ‘Social Groups’.
Complete List of Open Source Models:
New Healthcare models and Features
Integrates the incredible Spark NLP for Healthcare releases 3.3.4, 3.3.2 and 3.3.1, featuring:
- New Clinical NER Models for protected health information(PHI),
ner_biomarkerfor extracting extract biomarkers, therapies, oncological, and other general concepts- Oncogenes, Tumor_Finding, UnspecificTherapy, Ethnicity, Age, ResponseToTreatment, Biomarker, HormonalTherapy, Staging, Drug, CancerDx, Radiotherapy, CancerSurgery, TargetedTherapy, PerformanceStatus, CancerModifier, Radiological_Test_Result, Biomarker_Measurement, Metastasis, Radiological_Test, Chemotherapy, Test, Dosage, Test_Result, Immunotherapy, Date, Gender, Prognostic_Biomarkers, Duration, Predictive_Biomarkers
ner_nihss: NER model that can identify entities according to NIHSS guidelines for clinical stroke assessment to evaluate neurological status in acute stroke patients- 11_ExtinctionInattention, 6b_RightLeg, 1c_LOCCommands, 10_Dysarthria, NIHSS, 5_Motor, 8_Sensory, 4_FacialPalsy, 6_Motor, 2_BestGaze, Measurement, 6a_LeftLeg, 5b_RightArm, 5a_LeftArm, 1b_LOCQuestions, 3_Visual, 9_BestLanguage, 7_LimbAtaxia, 1a_LOC .
redl_nihss_biobert: relation extraction model that can relate scale items and their measurements according to NIHSS guidelines.es.med_ner.roberta_ner_diag_proc: New Spanish Clinical NER Models for extracting the entities DIAGNOSTICO, PROCEDIMIENTOes.resolve.snomed: New Spanish SNOMED Entity Resolversbert_sequence_classifier_question_statement_clinical:New Clinical Question vs Statement for BertForSequenceClassification modelmed_ner.covid_trials: This model is trained to extract covid-specific medical entities in clinical trials. It supports the following entities ranging from virus type to trial design: Stage, Severity, Virus, Trial_Design, Trial_Phase, N_Patients, Institution, Statistical_Indicator, Section_Header, Cell_Type, Cellular_component, Viral_components, Physiological_reaction, Biological_molecules, Admission_Discharge, Age, BMI, Cerebrovascular_Disease, Date, Death_Entity, Diabetes, Disease_Syndrome_Disorder, Dosage, Drug_Ingredient, Employment, Frequency, Gender, Heart_Disease, Hypertension, Obesity, Pulse, Race_Ethnicity, Respiration, Route, Smoking, Time, Total_Cholesterol, Treatment, VS_Finding, Vaccine .med_ner.chemd: This model extract the names of chemical compounds and drugs in medical texts. The entities that can be detected are as follows : SYSTEMATIC, IDENTIFIERS, FORMULA, TRIVIAL, ABBREVIATION, FAMILY, MULTIPLE . For reference click here . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4331685/bert_token_classifier_ner_bionlp: This model is BERT-based version of ner_bionlp model and can detect biological and genetics terms in cancer-related texts. (Amino_acid, Anatomical_system, Cancer, Cell, Cellular_component, Developing_anatomical_Structure, Gene_or_gene_product, Immaterial_anatomical_entity, Multi-tissue_structure, Organ, Organism, Organism_subdivision, Simple_chemical, Tissuebert_token_classifier_ner_cellular: This model is BERT-based version of ner_cellular model and can detect molecular biology-related terms (DNA, Cell_type, Cell_line, RNA, Protein) in medical texts.- We have updated
med_ner.jsl.enrichedmodel by enriching the training data using clinical trials data to make it more robust. This model is capable of predicting up to 87 different entities and is based on ner_jsl model. Here are the entities this model can detect; Social_History_Header, Oncology_Therapy, Blood_Pressure, Respiration, Performance_Status, Family_History_Header, Dosage, Clinical_Dept, Diet, Procedure, HDL, Weight, Admission_Discharge, LDL, Kidney_Disease, Oncological, Route, Imaging_Technique, Puerperium, Overweight, Temperature, Diabetes, Vaccine, Age, Test_Result, Employment, Time, Obesity, EKG_Findings, Pregnancy, Communicable_Disease, BMI, Strength, Tumor_Finding, Section_Header, RelativeDate, ImagingFindings, Death_Entity, Date, Cerebrovascular_Disease, Treatment, Labour_Delivery, Pregnancy_Delivery_Puerperium, Direction, Internal_organ_or_component, Psychological_Condition, Form, Medical_Device, Test, Symptom, Disease_Syndrome_Disorder, Staging, Birth_Entity, Hyperlipidemia, O2_Saturation, Frequency, External_body_part_or_region, Drug_Ingredient, Vital_Signs_Header, Substance_Quantity, Race_Ethnicity, VS_Finding, Injury_or_Poisoning, Medical_History_Header, Alcohol, Triglycerides, Total_Cholesterol, Sexually_Active_or_Sexual_Orientation, Female_Reproductive_Status, Relationship_Status, Drug_BrandName, RelativeTime, Duration, Hypertension, Metastasis, Gender, Oxygen_Therapy, Pulse, Heart_Disease, Modifier, Allergen, Smoking, Substance, Cancer_Modifier, Fetus_NewBorn, Height classify.bert_sequence.question_statement_clinical: This model classifies sentences into one of these two classes: question (interrogative sentence) or statement (declarative sentence) and trained with BertForSequenceClassification. This model is at first trained on SQuAD and SPAADIA dataset and then fine tuned on the clinical visit documents and MIMIC-III dataset annotated in-house. Using this model, you can find the question statements and exclude & utilize in the downstream tasks such as NER and relation extraction models.classify.token_bert.ner_chemical: This model is BERT-based version of ner_chemicals model and can detect chemical compounds (CHEM) in the medical texts.resolve.umls_disease_syndrome: This model is trained on the Disease or Syndrome category using sbiobert_base_cased_mli embeddings.
Complete List of Healthcare Models :
NLU Version 3.3.0
2000%+ Speedup on small data, 63 new models for 100+ Languages with 6 new supported Transformer classes including BERT, XLM-RoBERTa, alBERT, Longformer, XLnet based models, 48 NER profiling healthcare pipelines and much more in John Snow Labs NLU 3.3.0
We are incredibly excited to announce NLU 3.3.0 has been released!
It comes with a up to 2000%+ speedup on small datasets, 6 new Types of Deep Learning transformer models, including
RoBertaForTokenClassification,XlmRoBertaForTokenClassification,AlbertForTokenClassification,LongformerForTokenClassification,XlnetForTokenClassification,XlmRoBertaSentenceEmbeddings.
In total there are 63 NLP Models 6 New Languages Supported which are Igbo, Ganda, Dholuo, Naija, Wolof,Kinyarwanda with their corresponding ISO codes ig, lg, lou, pcm, wo,rw
with New SOTA XLM-RoBERTa models in Luganda, Kinyarwanda, Igbo, Hausa, and Amharic languages and 2 new Multilingual Embeddings with 100+ supported languages via XLM-Roberta are available.
On the healthcare NLP side we are glad to announce 18 new NLP for Healthcare models including
NER Profiling pretrained pipelines to run 48 different Clinical NER and 21 Different Biobert Models At Once Over the Input Text
New BERT-Based Deidentification NER Model, Sentence Entity Resolver Models For German Language
New Spell Checker Model For Drugs , 3 New Sentence Entity Resolver Models (3-char ICD10CM, RxNorm_NDC, HCPCS)
5 New Clinical NER Models (Trained By BertForTokenClassification Approach)
,Radiology NER Model Trained On cheXpert Datasetand New UMLS Sentence Entity Resolver Models
Additionally 2 new tutorials are avaiable, NLU & Streamlit Crashcourse and NLU for Healthcare Crashcourse of every of the 50 + healthcare Domains and 200+ healthcare models
New Features and Improvements
2000%+ Speedup prediction for small datasets
NLU pipelines now predict up to 2000% faster by optimizing integration with Spark NLP’s light pipelines.
NLU will configure usage of this automatically, but it can be turned off as well via multithread=False
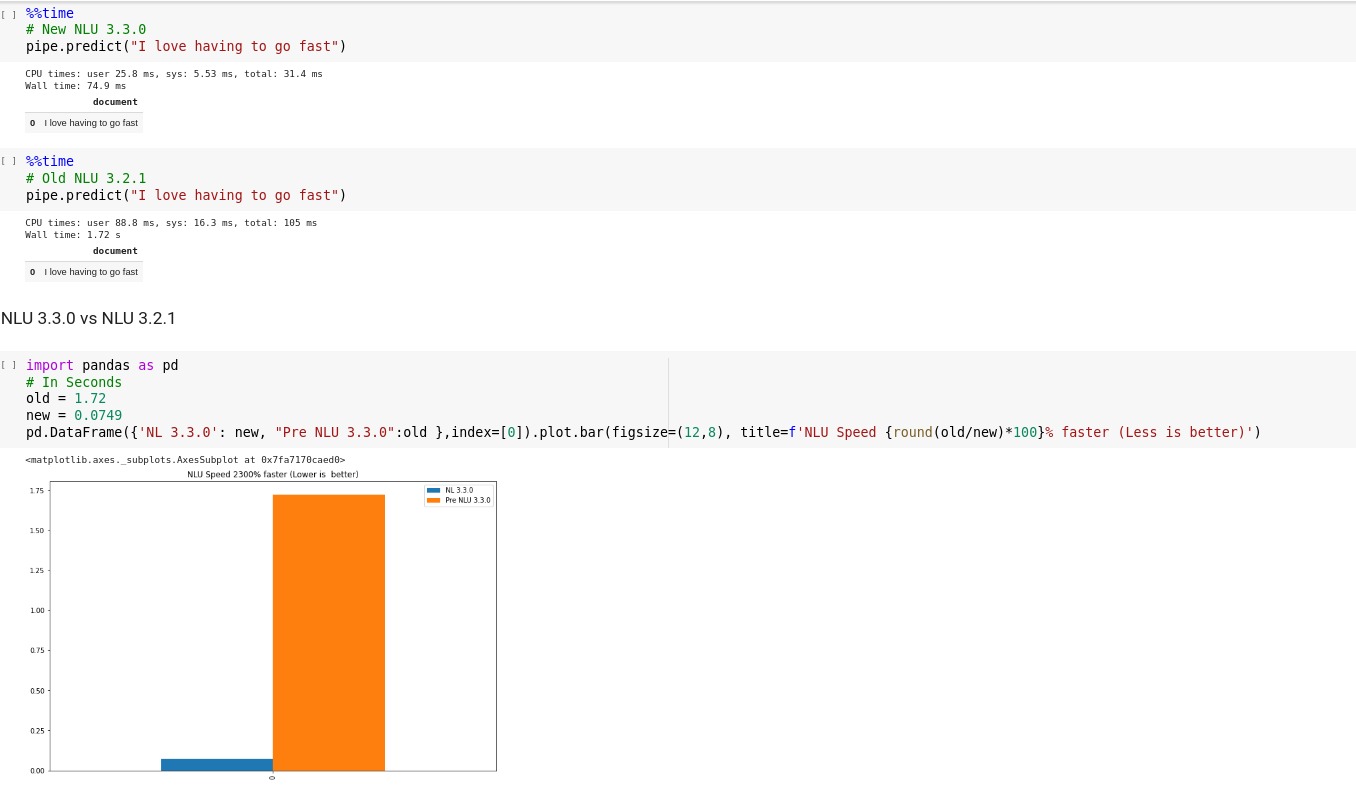
50x faster saving of NLU Pipelines
Up to 50x faster saving Spark NLP/ NLU models and pipelines! We have improved the way we package TensorFlow SavedModel while saving Spark NLP models & pipelines. For instance, it used to take up to 10 minutes to save the xlm_roberta_base model before Spark NLP 3.3.0, and now it only takes up to 15 seconds!
New Annotator Classes Integrated
The following new transformer classes are available with various pretrained weights in 1 line of code :
New Transformer Models
The following models are available from the amazing Spark NLP 3.3.0 and 3.3.1 releases which includes NLP models for Yiddish, Ukrainian, Telugu, Tamil, Somali, Sindhi, Russian, Punjabi, Nepali, Marathi, Malayalam, Kannada, Indonesian, Gujrati, Bosnian, Igbo, Ganda, Dholuo, Naija, Wolof,Kinyarwanda
New Healthcare models
The following models are available from the amazing Spark NLP for Healthcare releases 3.3.0, 3.2.3, 3.3.1, which includes 48 Multi-NER tuning pipelines, BERT-based DEidentification, German NER resolvers, Spell Checkers for Drugs, 5 ner NER models trained via BErtForTokenClassification, NER models for Radiology CID10CM, RxNORM NDC and HCPCSS models and UMLS sentence resolver models
Updated Model Names
The nlu model references have been updated to better reflect their use-cases.
- en.classify.token_bert.conll03
- en.classify.token_bert.large_conll03
- en.classify.token_bert.ontonote
- en.classify.token_bert.large_ontonote
- en.classify.token_bert.few_nerd
- en.classify.token_bert.classifier_ner_btc
- es.classify.token_bert.spanish_ner
- ja.classify.token_bert.classifier_ner_ud_gsd
- fa.classify.token_bert.parsbert_armanner
- fa.classify.token_bert.parsbert_ner
- fa.classify.token_bert.parsbert_peymaner
- sv.classify.token_bert.swedish_ner
- tr.classify.token_bert.turkish_ner
- en.classify.token_bert.ner_clinical
- en.classify.token_bert.ner_jsl
New Tutorial Videos
- NLU & Streamlit Crashcourse
- NLU for Healthcare Crashcourse of every of the 50 + healthcare Domains and 200+ healthcare models
Optional get_embeddings parameter for pipelines
NLU pipelines can now be forced to not return embeddings via get_embeddings parameter.
Updated Compatibility Docs
Added documentation section regarding compatibility of NLU, Spark NLP and Spark NLP for healthcare
Bugfixes
- Fixed a bug with Pyspark versions 3.0 and below that caused failure of predicting with pipeline
- Fixed a bug that caused the results of TokenClassifier Models to not be properly extracted
Additional NLU ressources
- 140+ NLU Tutorials
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.2.1
27 new models in 7 Languages, including Japanese NER, resolution models for SNOMED, ICDO, CPT and RxNorm codes and much more in NLU 3.2.1
We are very excited to announce NLU 3.2.1!
This release comes with models 27 new models for 7 languages which are transformer based.
New NER-Classifiers, BertSentenceEmbeddings, BertEmbeddings and BertForTokenClassificationEmbeddings
for Japanese, German, Dutch, Swedish, Spanish, French and English.
For healthcare there are new Entity Resolvers and MedicalNerModels
for Snomed Conditions, Cpt Measurements, Icd0, Rxnorm Dispositions, Posology and Deidentification.
Finally, a new tutorial notebook and a webinar are available, which showcase almost every feature of NLU
for the over 50 Domains in Healthcare/Clinical/Biomedical/etc..
New Transformer Models
Models in Japanese, German, Dutch, Swedish, Spanish, French and English from the great Spark NLP 3.2.3 release
New Healthcare Transformer Models
Models for Snomed Conditions, Cpt Measurements, Icd0, Rxnorm Dispositions, Posology and Deidentification from the amazing Spark NLP 3.2.2 for Healthcare Release
| nlu.load() Refrences | Spark NLP Refrence | Annotater class | Language |
|---|---|---|---|
| en.resolve.snomed_conditions | sbertresolve_snomed_conditions | SentenceEntityResolverModel | en |
| en.resolve.cpt.procedures_measurements | sbiobertresolve_cpt_procedures_measurements_augmented | SentenceEntityResolverModel | en |
| en.resolve.icdo.base | sbiobertresolve_icdo_base | SentenceEntityResolverModel | en |
| en.resolve.rxnorm.disposition.sbert | sbertresolve_rxnorm_disposition | SentenceEntityResolverModel | en |
| en.resolve.rxnorm_disposition.sbert | sbertresolve_rxnorm_disposition | SentenceEntityResolverModel | en |
| en.med_ner.posology.experimental | ner_posology_experimental | MedicalNerModel | en |
| en.med_ner.deid.subentity_augmented | ner_deid_subentity_augmented | MedicalNerModel | en |
New Notebooks
Enhancements
- Columns of the Pandas DataFrame returned by NLU will now be sorted alphabetically
Bugfixes
- Fixed a bug that caused output levels no beeing inferred properly
- Fixed a bug that caused SentenceResolver visualizations not to appear.
NLU Version 3.2.0
100+ Transformers Models in 40+ languages, 3-D Streamlit Entity-Embedding-Manifold visualizations, Multi-Lingual NER, Longformers, TokenDistilBERT, Trainable Sentence Resolvers, 7% less memory usage and much more in NLU 3.2.0
We are extremely excited to announce the release of NLU 3.2.0
which marks the 1-year anniversary of the birth of this magical library.
This release packs features and improvements in every division of NLU’s aspects,
89 new NLP models with new Models including Longformer, TokenBert, TokenDistilBert and Multi-Lingual NER for 40+ Languages.
12 new Healthcare models with trainable sentence resolvers and models Adverse Drug Relations, Clinical Token Bert Models, NER Models for Radiology, Drugs, Posology, Administration Cycles, RXNorm, and new Medical Assertion models.
New Streamlit visualizations enable you to see Entities in 3-D, 2-D, and 1-D Manifolds which are applicable to Entities and their Embeddings, Detected by Named-Entity-Recognizer models.
Finally, a ~7% decrease in Memory consumption in NLU’s core which benefits every computation, achieved by leveraging Pyarrow.
We are incredibly thankful to our community, which helped us come this far, and are looking forward to another magical year of NLU!
Streamlit Entity Manifold visualization
function pipe.viz_streamlit_entity_embed_manifold
Visualize recognized entities by NER models via their Entity Embeddings in 1-D, 2-D, or 3-D by Reducing Dimensionality via 10+ Supported methods from Manifold Algorithms
and Matrix Decomposition Algorithms.
You can pick additional NER models and compare them via the GUI dropdown on the left.
- Reduces Dimensionality of high dimensional Entity Embeddings to
1-D,2-D, or3-Dand plot the resulting data in an interactivePlotlyplot - Applicable with any of the 330+ Named Entity Recognizer models
- Gemerates
NUM-DIMENSIONS*NUM-NER-MODELS*NUM-DIMENSION-REDUCTION-ALGOSplots
nlu.load('ner').viz_streamlit_sentence_embed_manifold(['Hello From John Snow Labs', 'Peter loves to visit New York'])
or just run
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/09_entity_embedding_manifolds.py

function parameters pipe.viz_streamlit_sentence_embed_manifold
| Argument | Type | Default | Description | |
|---|---|---|---|---|
default_texts |
List[str] |
“Donald Trump likes to visit New York”, “Angela Merkel likes to visit Berlin!”, ‘Peter hates visiting Paris’) | List of strings to apply classifiers, embeddings, and manifolds to. | |
title |
str |
'NLU ❤️ Streamlit - Prototype your NLP startup in 0 lines of code🚀' |
Title of the Streamlit app | |
sub_title |
Optional[str] |
“Apply any of the 10+ Manifold or Matrix Decomposition algorithms to reduce the dimensionality of Entity Embeddings to 1-D, 2-D and 3-D “ |
Sub title of the Streamlit app | |
default_algos_to_apply |
List[str] |
["TSNE", "PCA"] |
A list Manifold and Matrix Decomposition Algorithms to apply. Can be either 'TSNE','ISOMAP','LLE','Spectral Embedding', 'MDS','PCA','SVD aka LSA','DictionaryLearning','FactorAnalysis','FastICA' or 'KernelPCA', |
|
target_dimensions |
List[int] |
(1,2,3) |
Defines the target dimension embeddings will be reduced to | |
show_algo_select |
bool |
True |
Show selector for Manifold and Matrix Decomposition Algorithms | |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. | |
num_cols |
int |
2 |
How many columns should for the layout in streamlit when rendering the similarity matrixes. | |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn | |
show_logo |
bool |
True |
Show logo | |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. | |
n_jobs |
Optional[int] |
3 |
False |
How many cores to use for paralellzing when using Sklearn Dimension Reduction algorithms. |
Sentence Entity Resolver Training
Sentence Entity Resolver Training Tutorial Notebook
Named Entities are sub pieces in textual data which are labeled with classes.
These classes and strings are still ambiguous though and it is not possible to group semantically identically entities without any definition of terminology.
With the Sentence Resolver you can train a state-of-the-art deep learning architecture to map entities to their unique terminological representation.
Train a Sentence resolver on a dataset with columns named y , _y and text. y is a label, _y is an extra identifier label, text is the raw text
import pandas as pd
import nlu
dataset = pd.DataFrame({
'text': ['The Tesla company is good to invest is', 'TSLA is good to invest','TESLA INC. we should buy','PUT ALL MONEY IN TSLA inc!!'],
'y': ['23','23','23','23'],
'_y': ['TESLA','TESLA','TESLA','TESLA'],
})
trainable_pipe = nlu.load('train.resolve_sentence')
fitted_pipe = trainable_pipe.fit(dataset)
res = fitted_pipe.predict(dataset)
fitted_pipe.predict(["Peter told me to buy Tesla ", 'I have money to loose, is TSLA a good option?'])
| sentence_resolution_resolve_sentence_confidence | sentence_resolution_resolve_sentence_code | sentence_resolution_resolve_sentence | sentence | |
|---|---|---|---|---|
| 0 | ‘1.0000’ | ‘23’ | ‘TESLA’ | ‘The Tesla company is good to invest is’ |
| 1 | ‘1.0000’ | ‘23’ | ‘TESLA’ | ‘TSLA is good to invest’ |
| 2 | ‘1.0000’ | ‘23’ | ‘TESLA’ | ‘TESLA INC. we should buy’ |
| 3 | ‘1.0000’ | ‘23’ | ‘TESLA’ | ‘PUT ALL MONEY IN TSLA inc!!’ |
Alternatively you can also use non-default healthcare embeddings.
trainable_pipe = nlu.load('en.embed.glove.biovec train.resolve_sentence')
Transformer Models
New models from the spectacular Spark NLP 3.2.0 + releases are integrated.
89 new models in total, with new LongFormer, TokenBert, TokenDistilBert and Multi-Lingual NER for 40+ languages.
The supported languages with their ISO 639-1 code are : af, ar, bg, bn, de, el, en, es, et, eu, fa, fi, fr, he, hi, hu, id, it, ja, jv, ka, kk, ko, ml, mr, ms, my, nl, pt, ru, sw, ta, te, th, tl, tr, ur, vi, yo, and zh
New Healthcare Transformer Models
12 new models from the amazing Spark NLP for Healthcare 3.2.0+ releases, including models for genetic variants, radiology, assertion,
rxnorm, adverse drugs and new clinical tokenbert models that improves accuracy by 4% compared to the previous models.
| nlu.load() Refrence | Spark NLP Refrence | Annotator Class |
|---|---|---|
| en.med_ner.radiology.wip_greedy_biobert | jsl_rd_ner_wip_greedy_biobert | MedicalNerModel |
| en.med_ner.genetic_variants | ner_genetic_variants | MedicalNerModel |
| en.med_ner.jsl_slim | ner_jsl_slim | MedicalNerModel |
| en.med_ner.jsl_greedy_biobert | ner_jsl_greedy_biobert | MedicalNerModel |
| en.embed.token_bert.ner_clinical | bert_token_classifier_ner_clinical | MedicalNerModel |
| en.embed.token_bert.ner_jsl | bert_token_classifier_ner_jsl | MedicalNerModel |
| en.relation.ade | redl_ade_biobert | RelationExtractionDLModel |
| en.relation.ade_clinical | re_ade_clinical | RelationExtractionDLModel |
| en.relation.ade_biobert | re_ade_biobert | RelationExtractionDLModel |
| en.resolve.rxnorm_disposition | sbiobertresolve_rxnorm_disposition | SentenceEntityResolverModel |
| en.assert.jsl | assertion_jsl | AssertionDLModel |
| en.assert.jsl_large | assertion_jsl_large | AssertionDLModel |
PyArrow Memory Optimizations
Optimized integration with Pyarrow to share memory between the Python Virtual Machine and Java Virtual Machine which yields around
7% less memory consumption on average in all computations. This improvement will take effect for everyone using the default pyspark installation, which comes with a compatible Pyarrow Version.
If you manually install or upgrade Pyarrow, please refer to the official Spark docs and make sure
you have a Pyarrow version installed that works with your Pyspark version.

New Notebooks
Bugfixes
- Fixed a bug that caused the similarity matrix calculations to generate NaNs and crash
Additional NLU ressources
- 140+ NLU Tutorials
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark streamlit==0.80.0`
NLU Version 3.1.1
Sentence Embedding Visualizations, 20+ New Models, 2 New Trainable Models, Drug Normalizer and more in John Snow Labs NLU 3.1.1
We are very excited to announce NLU 3.1.1 has been released!
It features a new Sentence Embedding visualization component for Streamlit which supports all 10+ previous dimension
reduction techniques. Additionally, all embedding visualizations now support Latent Dirichlet Allocation for dimension reduction.
Finally, 2 new trainable models for NER and chunk resolution are supported, a new drug normalizer algorithm has been added,
20+ new pre-trained models including Multi-Lingual, German,
various healthcare models and improved NER defaults when using licensed models that have NER dependencies.
Streamlit Sentence Embedding visualization via Manifold and Matrix Decomposition algorithms
function pipe.viz_streamlit_sentence_embed_manifold
Visualize Sentence Embeddings in 1-D, 2-D, or 3-D by Reducing Dimensionality via 12 Supported methods from Manifold Algorithms
and Matrix Decomposition Algorithms.
Additionally, you can color the lower dimensional points with a label that has been previously assigned to the text by specifying a list of nlu references in the additional_classifiers_for_coloring parameter.
You can also select additional classifiers via the GUI.
- Reduces Dimensionality of high dimensional Sentence Embeddings to
1-D,2-D, or3-Dand plot the resulting data in an interactivePlotlyplot - Applicable with any of the 100+ Sentence Embedding models
- Color points by classifying with any of the 100+ Document Classifiers
- Gemerates
NUM-DIMENSIONS*NUM-EMBEDDINGS*NUM-DIMENSION-REDUCTION-ALGOSplots
text= """You can visualize any of the 100 + Sentence Embeddings
with 10+ dimension reduction algorithms
and view the results in 3D, 2D, and 1D
which can be colored by various classifier labels!
"""
nlu.load('embed_sentence.bert').viz_streamlit_sentence_embed_manifold(text)

function parameters pipe.viz_streamlit_sentence_embed_manifold
| Argument | Type | Default | Description | |
|---|---|---|---|---|
default_texts |
List[str] |
(“Donald Trump likes to party!”, “Angela Merkel likes to party!”, ‘Peter HATES TO PARTTY!!!! :(‘) | List of strings to apply classifiers, embeddings, and manifolds to. | |
text |
Optional[str] |
'Billy likes to swim' |
Text to predict classes for. | |
sub_title |
Optional[str] |
“Apply any of the 11 Manifold or Matrix Decomposition algorithms to reduce the dimensionality of Sentence Embeddings to 1-D, 2-D and 3-D “ |
Sub title of the Streamlit app | |
default_algos_to_apply |
List[str] |
["TSNE", "PCA"] |
A list Manifold and Matrix Decomposition Algorithms to apply. Can be either 'TSNE','ISOMAP','LLE','Spectral Embedding', 'MDS','PCA','SVD aka LSA','DictionaryLearning','FactorAnalysis','FastICA' or 'KernelPCA', |
|
target_dimensions |
List[int] |
(1,2,3) |
Defines the target dimension embeddings will be reduced to | |
show_algo_select |
bool |
True |
Show selector for Manifold and Matrix Decomposition Algorithms | |
show_embed_select |
bool |
True |
Show selector for Embedding Selection | |
show_color_select |
bool |
True |
Show selector for coloring plots | |
display_embed_information |
bool |
True |
Show additional embedding information like dimension, nlu_reference, spark_nlp_reference, sotrage_reference, modelhub link and more. |
|
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. | |
num_cols |
int |
2 |
How many columns should for the layout in streamlit when rendering the similarity matrixes. | |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn | |
additional_classifiers_for_coloring |
List[str] |
['sentiment.imdb'] |
List of additional NLU references to load for generting hue colors | |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click | |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
|
show_logo |
bool |
True |
Show logo | |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. | |
n_jobs |
Optional[int] |
3 |
False |
How many cores to use for paralellzing when using Sklearn Dimension Reduction algorithms. |
General Streamlit enhancements
Support for Latent Dirichlet Allocation
The Latent Dirichlet Allocation algorithm is now supported for the Word Embedding Visualizations and the Sentence Embedding Visualizations.
Normalization of Vectors before calculating sentence similarity.
WordEmbedding vectors will now be normalized before calculating similarity scores, which bounds each similarity between 0 and 1
Control order of plots
You can now control the order in Which visualizations appear in the main GUI
Sentence Embedding Visualization
Chunk Entity Resolver Training
Chunk Entity Resolver Training Tutorial Notebook
Named Entities are sub pieces in textual data which are labeled with classes.
These classes and strings are still ambigous though and it is not possible to group semantically identically entities without any definition of terminology.
With the Chunk Resolver you can train a state-of-the-art deep learning architecture to map entities to their unique terminological representation.
Train a chunk resolver on a dataset with columns named y , _y and text. y is a label, _y is an extra identifier label, text is the raw text
import pandas as pd
dataset = pd.DataFrame({
'text': ['The Tesla company is good to invest is', 'TSLA is good to invest','TESLA INC. we should buy','PUT ALL MONEY IN TSLA inc!!'],
'y': ['23','23','23','23']
'_y': ['TESLA','TESLA','TESLA','TESLA'],
})
trainable_pipe = nlu.load('train.resolve_chunks')
fitted_pipe = trainable_pipe.fit(dataset)
res = fitted_pipe.predict(dataset)
fitted_pipe.predict(["Peter told me to buy Tesla ", 'I have money to loose, is TSLA a good option?'])
| entity_resolution_confidence | entity_resolution_code | entity_resolution | document |
|---|---|---|---|
| ‘1.0000’ | ‘23’ | ‘TESLA’ | Peter told me to buy Tesla |
| ‘1.0000’ | ‘23’ | ‘TESLA’ | I have money to loose, is TSLA a good option? |
Train with default glove embeddings
untrained_chunk_resolver = nlu.load('train.resolve_chunks')
trained_chunk_resolver = untrained_chunk_resolver.fit(df)
trained_chunk_resolver.predict(df)
Train with custom embeddings
# Use BIo GLove
untrained_chunk_resolver = nlu.load('en.embed.glove.biovec train.resolve_chunks')
trained_chunk_resolver = untrained_chunk_resolver.fit(df)
trained_chunk_resolver.predict(df)
Rule based NER with Context Matcher
Rule based NER with context matching tutorial notebook
Define a rule-based NER algorithm by providing Regex Patterns and resolution mappings.
The confidence value is computed using a heuristic approach based on how many matches it has.
A dictionary can be provided with setDictionary to map extracted entities to a unified representation. The first column of the dictionary file should be the representation with the following columns the possible matches.
import nlu
import json
# Define helper functions to write NER rules to file
"""Generate json with dict contexts at target path"""
def dump_dict_to_json_file(dict, path):
with open(path, 'w') as f: json.dump(dict, f)
"""Dump raw text file """
def dump_file_to_csv(data,path):
with open(path, 'w') as f:f.write(data)
sample_text = """A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and subsequent type two diabetes mellitus ( T2DM ), one prior episode of HTG-induced pancreatitis three years prior to presentation , associated with an acute hepatitis , and obesity with a body mass index ( BMI ) of 33.5 kg/m2 , presented with a one-week history of polyuria , polydipsia , poor appetite , and vomiting. Two weeks prior to presentation , she was treated with a five-day course of amoxicillin for a respiratory tract infection . She was on metformin , glipizide , and dapagliflozin for T2DM and atorvastatin and gemfibrozil for HTG . She had been on dapagliflozin for six months at the time of presentation . Physical examination on presentation was significant for dry oral mucosa ; significantly , her abdominal examination was benign with no tenderness , guarding , or rigidity . Pertinent laboratory findings on admission were : serum glucose 111 mg/dl , bicarbonate 18 mmol/l , anion gap 20 , creatinine 0.4 mg/dL , triglycerides 508 mg/dL , total cholesterol 122 mg/dL , glycated hemoglobin ( HbA1c ) 10% , and venous pH 7.27 . Serum lipase was normal at 43 U/L . Serum acetone levels could not be assessed as blood samples kept hemolyzing due to significant lipemia . The patient was initially admitted for starvation ketosis , as she reported poor oral intake for three days prior to admission . However , serum chemistry obtained six hours after presentation revealed her glucose was 186 mg/dL , the anion gap was still elevated at 21 , serum bicarbonate was 16 mmol/L , triglyceride level peaked at 2050 mg/dL , and lipase was 52 U/L . β-hydroxybutyrate level was obtained and found to be elevated at 5.29 mmol/L - the original sample was centrifuged and the chylomicron layer removed prior to analysis due to interference from turbidity caused by lipemia again . The patient was treated with an insulin drip for euDKA and HTG with a reduction in the anion gap to 13 and triglycerides to 1400 mg/dL , within 24 hours . Twenty days ago. Her euDKA was thought to be precipitated by her respiratory tract infection in the setting of SGLT2 inhibitor use . At birth the typical boy is growing slightly faster than the typical girl, but the velocities become equal at about seven months, and then the girl grows faster until four years. From then until adolescence no differences in velocity can be detected. 21-02-2020 21/04/2020 """
# Define Gender NER matching rules
gender_rules = {
"entity": "Gender",
"ruleScope": "sentence",
"completeMatchRegex": "true" }
# Define dict data in csv format
gender_data = '''male,man,male,boy,gentleman,he,him
female,woman,female,girl,lady,old-lady,she,her
neutral,neutral'''
# Dump configs to file
dump_dict_to_json_file(gender_data, 'gender.csv')
dump_dict_to_json_file(gender_rules, 'gender.json')
gender_NER_pipe = nlu.load('match.context')
gender_NER_pipe.print_info()
gender_NER_pipe['context_matcher'].setJsonPath('gender.json')
gender_NER_pipe['context_matcher'].setDictionary('gender.csv', options={"delimiter":","})
gender_NER_pipe.predict(sample_text)
| context_match | context_match_confidence |
|---|---|
| female | 0.13 |
| she | 0.13 |
| she | 0.13 |
| she | 0.13 |
| she | 0.13 |
| boy | 0.13 |
| girl | 0.13 |
| girl | 0.13 |
Context Matcher Parameters
You can define the following parameters in your rules.json file to define the entities to be matched
| Parameter | Type | Description |
|---|---|---|
| entity | str |
The name of this rule |
| regex | Optional[str] |
Regex Pattern to extract candidates |
| contextLength | Optional[int] |
defines the maximum distance a prefix and suffix words can be away from the word to match,whereas context are words that must be immediately after or before the word to match |
| prefix | Optional[List[str]] |
Words preceding the regex match, that are at most contextLength characters aways |
| regexPrefix | Optional[str] |
RegexPattern of words preceding the regex match, that are at most contextLength characters aways |
| suffix | Optional[List[str]] |
Words following the regex match, that are at most contextLength characters aways |
| regexSuffix | Optional[str] |
RegexPattern of words following the regex match, that are at most contextLength distance aways |
| context | Optional[List[str]] |
list of words that must be immediatly before/after a match |
| contextException | Optional[List[str]] |
?? List of words that may not be immediatly before/after a match |
| exceptionDistance | Optional[int] |
Distance exceptions must be away from a match |
| regexContextException | Optional[str] |
Regex Pattern of exceptions that may not be within exceptionDistance range of the match |
| matchScope | Optional[str] |
Either token or sub-token to match on character basis |
| completeMatchRegex | Optional[str] |
Wether to use complete or partial matching, either "true" or "false" |
| ruleScope | str |
currently only sentence supported |
Drug Normalizer
Drug Normalizer tutorial notebook
Normalize raw text from clinical documents, e.g. scraped web pages or xml documents. Removes all dirty characters from text following one or more input regex patterns. Can apply unwanted character removal which a specific policy. Can apply lower case normalization.
Parameters are
- lowercase: whether to convert strings to lowercase. Default is False.
policy: rule to remove patterns from text. Valid policy values are:allabbreviations,dosagesDefaults isall.abbreviationpolicy used to expend common drugs abbreviations,dosagespolicy used to convert drugs dosages and values to the standard form (see examples below).
data = ["Agnogenic one half cup","adalimumab 54.5 + 43.2 gm","aspirin 10 meq/ 5 ml oral sol","interferon alfa-2b 10 million unit ( 1 ml ) injec","Sodium Chloride/Potassium Chloride 13bag"]
nlu.load('norm_drugs').predict(data)
| drug_norm | text |
|---|---|
| Agnogenic 0.5 oral solution | Agnogenic one half cup |
| adalimumab 97700 mg | adalimumab 54.5 + 43.2 gm |
| aspirin 2 meq/ml oral solution | aspirin 10 meq/ 5 ml oral sol |
| interferon alfa - 2b 10000000 unt ( 1 ml ) injection | interferon alfa-2b 10 million unit ( 1 ml ) injec |
| Sodium Chloride / Potassium Chloride 13 bag | Sodium Chloride/Potassium Chloride 13bag |
New NLU Spells
These new magical 1-liners which get new the folowing models
Open Source NLU Spells
| NLU Spell | Spark NLP Model |
|---|---|
| nlu.load(‘de.ner.wikiner.6B_100’) | wikiner_6B_100 |
| nlu.load(‘xx.embed.glove.glove_6B_100’) | glove_6B_100 |
Healthcare NLU spells
Improved NER defaults
When loading licensed models that require a NER features like Assertion, Relation, Resolution,
nlu will now use the en.med_ner model which maps to the Spark NLP model jsl_ner_wip_clinical as default.
See https://nlp.johnsnowlabs.com/2021/03/31/jsl_ner_wip_clinical_en.html for more infos on this model.
New Notebooks
Additional NLU ressources
- 140+ NLU Tutorials
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark==3.0.3
NLU Version 3.1.0
2600+ New Models for 200+ Languages and 10+ Dimension Reduction Algorithms for Streamlit Word-Embedding visualizations in 3-D
We are extremely excited to announce the release of NLU 3.1 !
This is our biggest release so far and it comes with over 2600+ new models in 200+ languages, including DistilBERT, RoBERTa, and XLM-RoBERTa and Huggingface based Embeddings from the incredible Spark-NLP 3.1.0 release,
new Streamlit Visualizations for visualizing Word Embeddings in 3-D, 2-D, and 1-D,
New Healthcare pipelines for healthcare code mappings
and finally confidence extraction for open source NER models.
Additionally, the NLU Namespace has been renamed to the NLU Spellbook, to reflect the magicalness of each 1-liners represented by them!
Streamlit Word Embedding visualization via Manifold and Matrix Decomposition algorithms
function pipe.viz_streamlit_word_embed_manifold
Visualize Word Embeddings in 1-D, 2-D, or 3-D by Reducing Dimensionality via 11 Supported methods from Manifold Algorithms
and Matrix Decomposition Algorithms.
Additionally, you can color the lower dimensional points with a label that has been previously assigned to the text by specifying a list of nlu references in the additional_classifiers_for_coloring parameter.
- Reduces Dimensionality of high dimensional Word Embeddings to
1-D,2-D, or3-Dand plot the resulting data in an interactivePlotlyplot - Applicable with any of the 100+ Word Embedding models
- Color points by classifying with any of the 100+ Parts of Speech Classifiers or Document Classifiers
- Gemerates
NUM-DIMENSIONS*NUM-EMBEDDINGS*NUM-DIMENSION-REDUCTION-ALGOSplots
nlu.load('bert',verbose=True).viz_streamlit_word_embed_manifold(default_texts=THE_MATRIX_ARCHITECT_SCRIPT.split('\n'),default_algos_to_apply=['TSNE'],MAX_DISPLAY_NUM=5)

function parameters pipe.viz_streamlit_word_embed_manifold
| Argument | Type | Default | Description |
|---|---|---|---|
default_texts |
List[str] |
(“Donald Trump likes to party!”, “Angela Merkel likes to party!”, ‘Peter HATES TO PARTTY!!!! :(‘) | List of strings to apply classifiers, embeddings, and manifolds to. |
text |
Optional[str] |
'Billy likes to swim' |
Text to predict classes for. |
sub_title |
Optional[str] |
Apply any of the 11 Manifold or Matrix Decomposition algorithms to reduce the dimensionality of Word Embeddings to 1-D, 2-D and 3-D |
Sub title of the Streamlit app |
default_algos_to_apply |
List[str] |
["TSNE", "PCA"] |
A list Manifold and Matrix Decomposition Algorithms to apply. Can be either 'TSNE','ISOMAP','LLE','Spectral Embedding', 'MDS','PCA','SVD aka LSA','DictionaryLearning','FactorAnalysis','FastICA' or 'KernelPCA' |
target_dimensions |
List[int] |
(1,2,3) |
Defines the target dimension embeddings will be reduced to |
show_algo_select |
bool |
True |
Show selector for Manifold and Matrix Decomposition Algorithms |
show_embed_select |
bool |
True |
Show selector for Embedding Selection |
show_color_select |
bool |
True |
Show selector for coloring plots |
MAX_DISPLAY_NUM |
int |
100 |
Cap maximum number of Tokens displayed |
display_embed_information |
bool |
True |
Show additional embedding information like dimension, nlu_reference, spark_nlp_reference, sotrage_reference, modelhub link and more. |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
num_cols |
int |
2 |
How many columns should for the layout in streamlit when rendering the similarity matrixes. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
additional_classifiers_for_coloring |
List[str] |
['pos', 'sentiment.imdb'] |
List of additional NLU references to load for generting hue colors |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
n_jobs |
Optional[int] |
False |
How many cores to use for paralellzing when using Sklearn Dimension Reduction algorithms. |
Larger Example showcasing more dimension reduction techniques on a larger corpus:

Supported Manifold Algorithms
New Healthcare Pipelines
Five new healthcare code mapping pipelines:
nlu.load(en.resolve.icd10cm.umls): This pretrained pipeline maps ICD10CM codes to UMLS codes without using any text data. You’ll just feed white space-delimited ICD10CM codes and it will return the corresponding UMLS codes as a list. If there is no mapping, the original code is returned with no mapping.
{'icd10cm': ['M89.50', 'R82.2', 'R09.01'],'umls': ['C4721411', 'C0159076', 'C0004044']}
nlu.load(en.resolve.mesh.umls): This pretrained pipeline maps MeSH codes to UMLS codes without using any text data. You’ll just feed white space-delimited MeSH codes and it will return the corresponding UMLS codes as a list. If there is no mapping, the original code is returned with no mapping.
{'mesh': ['C028491', 'D019326', 'C579867'],'umls': ['C0970275', 'C0886627', 'C3696376']}
nlu.load(en.resolve.rxnorm.umls): This pretrained pipeline maps RxNorm codes to UMLS codes without using any text data. You’ll just feed white space-delimited RxNorm codes and it will return the corresponding UMLS codes as a list. If there is no mapping, the original code is returned with no mapping.
{'rxnorm': ['1161611', '315677', '343663'],'umls': ['C3215948', 'C0984912', 'C1146501']}
nlu.load(en.resolve.rxnorm.mesh): This pretrained pipeline maps RxNorm codes to MeSH codes without using any text data. You’ll just feed white space-delimited RxNorm codes and it will return the corresponding MeSH codes as a list. If there is no mapping, the original code is returned with no mapping.
{'rxnorm': ['1191', '6809', '47613'],'mesh': ['D001241', 'D008687', 'D019355']}
nlu.load(en.resolve.snomed.umls): This pretrained pipeline maps SNOMED codes to UMLS codes without using any text data. You’ll just feed white space-delimited SNOMED codes and it will return the corresponding UMLS codes as a list. If there is no mapping, the original code is returned with no mapping.{'snomed': ['733187009', '449433008', '51264003'],'umls': ['C4546029', 'C3164619', 'C0271267']}
In the following table the NLU and Spark-NLP references are listed:
New Open Source Models and Pipelines
Bugfixes
-
Fixed bugs that occured when loading a model from disk.
- 140+ NLU Tutorials
- Streamlit visualizations docs
- The complete list of all 1100+ models & pipelines in 192+ languages is available on Models Hub.
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark==3.0.3
NLU Version 3.0.2
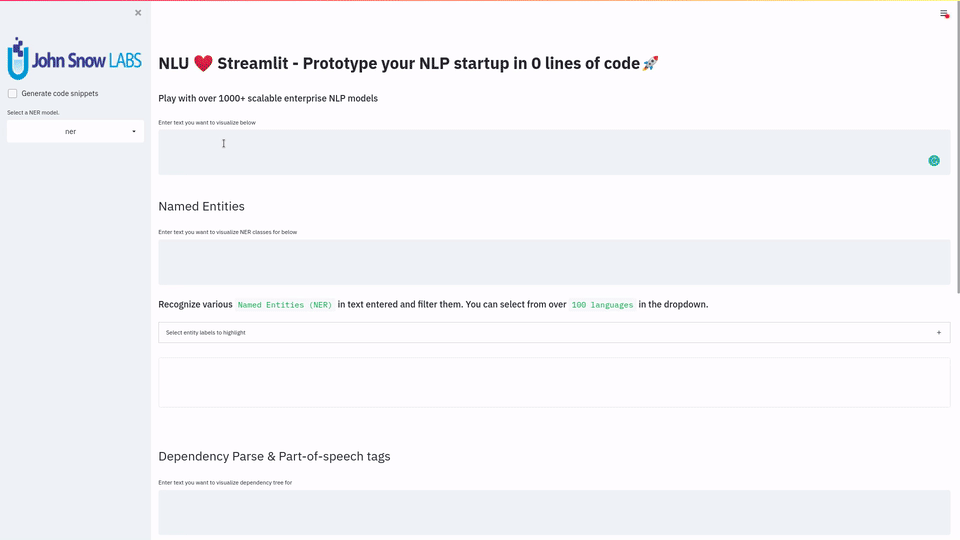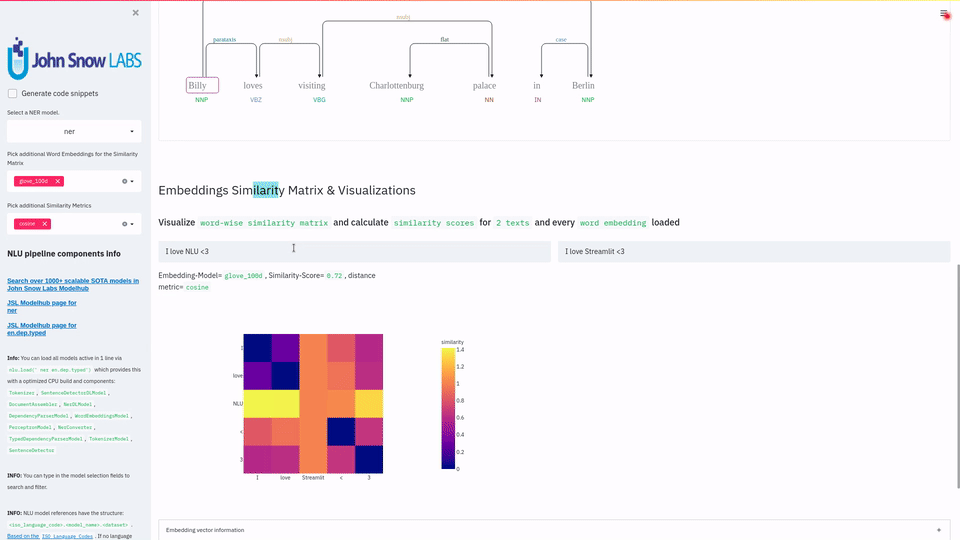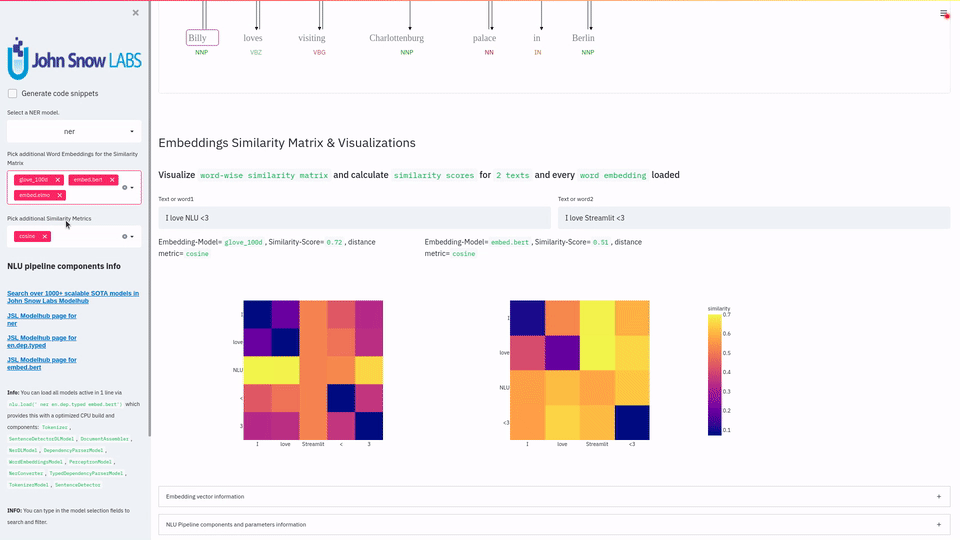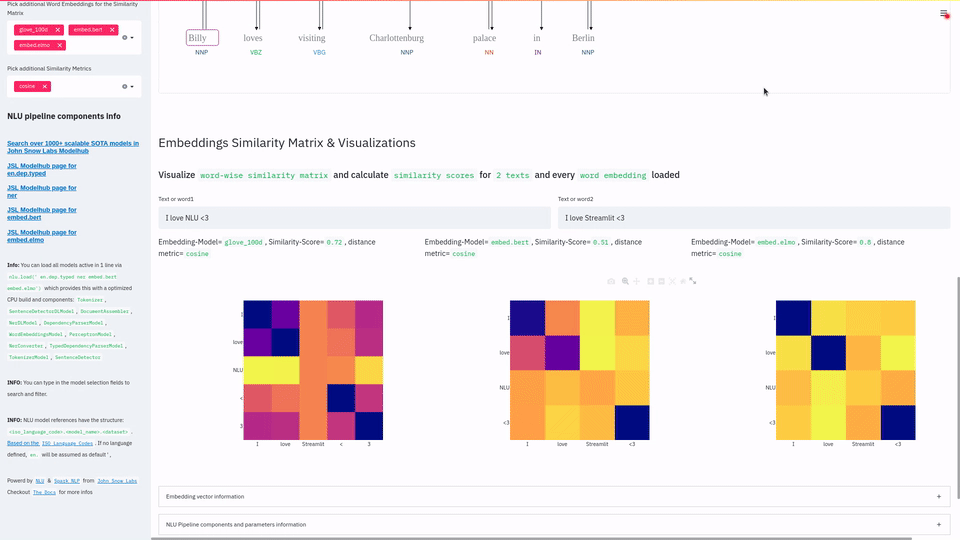
This release contains examples and tutorials on how to visualize the 1000+ state-of-the-art NLP models provided by NLU in just 1 line of code in streamlit.
It includes simple 1-liners you can sprinkle into your Streamlit app to for features like Dependency Trees, Named Entities (NER), text classification results, semantic simmilarity,
embedding visualizations via ELMO, BERT, ALBERT, XLNET and much more . Additionally, improvements for T5, various resolvers have been added and models Farsi, Hebrew, Korean, and Turkish
This is the ultimate NLP research tool. You can visualize and compare the results of hundreds of context aware deep learning embeddings and compare them with classical vanilla embeddings like Glove
and can see with your own eyes how context is encoded by transformer models like BERT or XLNETand many more !
Besides that, you can also compare the results of the 200+ NER models John Snow Labs provides and see how peformances changes with varrying ebeddings, like Contextual, Static and Domain Specific Embeddings.
Install
For detailed instructions refer to the NLU install documentation here
You need Open JDK 8 installed and the following python packages
pip install nlu streamlit pyspark==3.0.1 sklearn plotly
Problems? Connect with us on Slack!
Impatient and want some action?
Just run this Streamlit app, you can use it to generate python code for each NLU-Streamlit building block
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/01_dashboard.py
Quick Starter cheat sheet - All you need to know in 1 picture for NLU + Streamlit
For NLU models to load, see the NLU Namespace or the John Snow Labs Modelshub or go straight to the source.
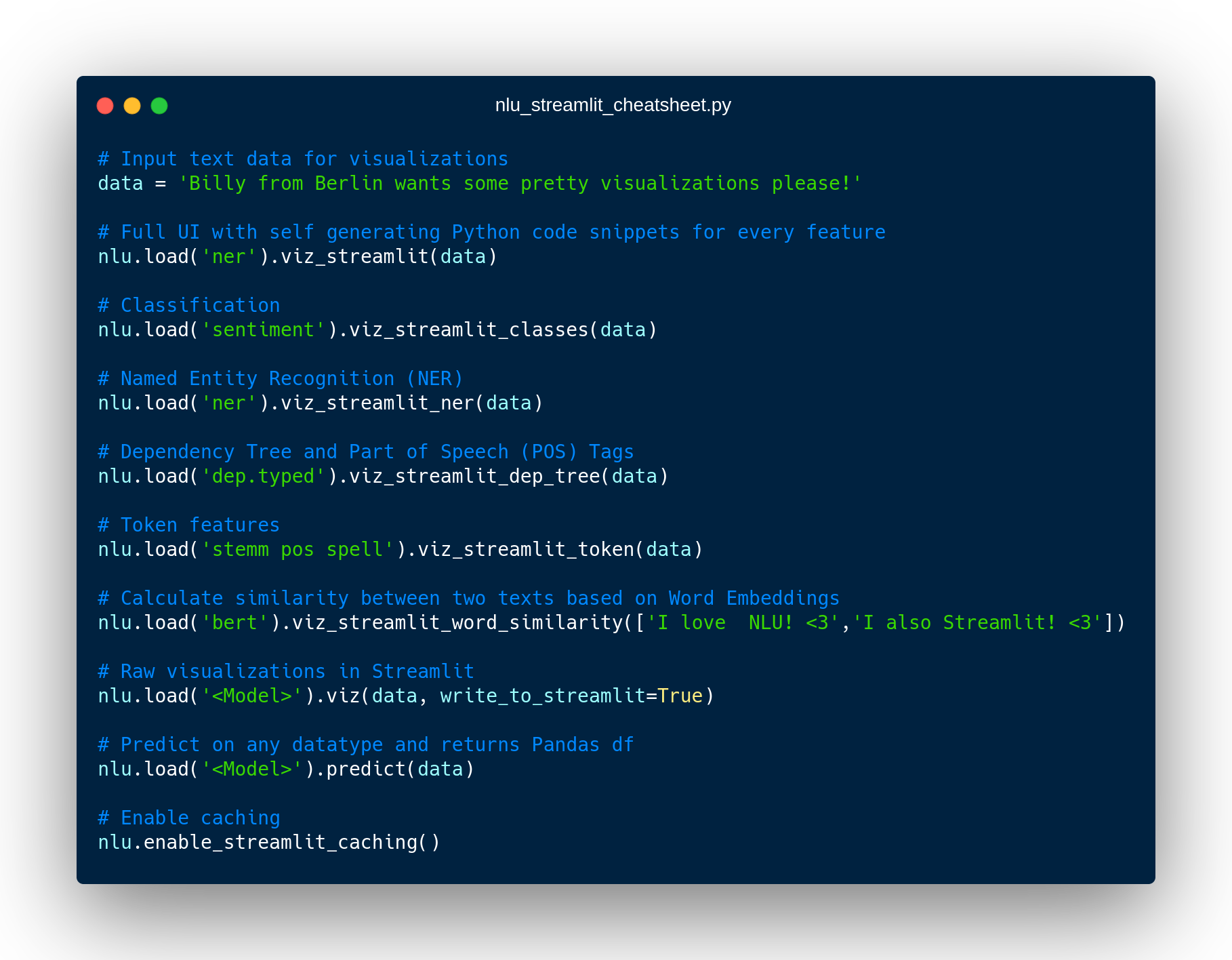
Examples
Just try out any of these. You can use the first example to generate python-code snippets which you can recycle as building blocks in your streamlit apps!
Example: 01_dashboard
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/01_dashboard.py
Example: 02_NER
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/02_NER.py
Example: 03_text_similarity_matrix
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/03_text_similarity_matrix.py
Example: 04_dependency_tree
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/04_dependency_tree.py
Example: 05_classifiers
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/05_classifiers.py
Example: 06_token_features
streamlit run https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/examples/streamlit/06_token_features.py
How to use NLU?
All you need to know about NLU is that there is the nlu.load() method which returns a NLUPipeline object
which has a .predict() that works on most common data types in the pydata stack like Pandas dataframes .
Ontop of that, there are various visualization methods a NLUPipeline provides easily integrate in Streamlit as re-usable components. viz() method
Overview of NLU + Streamlit buildingblocks
| Method | Description |
|---|---|
nlu.load('<Model>').predict(data) |
Load any of the 1000+ models by providing the model name any predict on most Pythontic data strucutres like Pandas, strings, arrays of strings and more |
nlu.load('<Model>').viz_streamlit(data) |
Display full NLU exploration dashboard, that showcases every feature avaiable with dropdown selectors for 1000+ models |
nlu.load('<Model>').viz_streamlit_similarity([string1, string2]) |
Display similarity matrix and scalar similarity for every word embedding loaded and 2 strings. |
nlu.load('<Model>').viz_streamlit_ner(data) |
Visualize predicted NER tags from Named Entity Recognizer model |
nlu.load('<Model>').viz_streamlit_dep_tree(data) |
Visualize Dependency Tree together with Part of Speech labels |
nlu.load('<Model>').viz_streamlit_classes(data) |
Display all extracted class features and confidences for every classifier loaded in pipeline |
nlu.load('<Model>').viz_streamlit_token(data) |
Display all detected token features and informations in Streamlit |
nlu.load('<Model>').viz(data, write_to_streamlit=True) |
Display the raw visualization without any UI elements. See viz docs for more info. By default all aplicable nlu model references will be shown. |
nlu.enable_streamlit_caching() |
Enable caching the nlu.load() call. Once enabled, the nlu.load() method will automatically cached. This is recommended to run first and for large peformance gans |
Detailed visualizer information and API docs
function pipe.viz_streamlit
Display a highly configurable UI that showcases almost every feature available for Streamlit visualization with model selection dropdowns in your applications.
Ths includes :
Similarity Matrix&Scalars&Embedding Informationfor any of the 100+ Word Embedding ModelsNER visualizationsfor any of the 200+ Named entity recognizersLabled&Unlabled Dependency Trees visualizationswithPart of Speech Tagsfor any of the 100+ Part of Speech ModelsToken informationspredicted by any of the 1000+ modelsClassification resultspredicted by any of the 100+ models classification modelsPipeline Configuration&Model Information&Link to John Snow Labs Modelshubfor all loaded pipelinesAuto generate Python codethat can be copy pasted to re-create the individual Streamlit visualization blocks. NlLU takes the first model specified asnlu.load()for the first visualization run.
Once the Streamlit app is running, additional models can easily be added via the UI.
It is recommended to run this first, since you can generate Python code snippetsto recreate individual Streamlit visualization blocks
nlu.load('ner').viz_streamlit(['I love NLU and Streamlit!','I hate buggy software'])
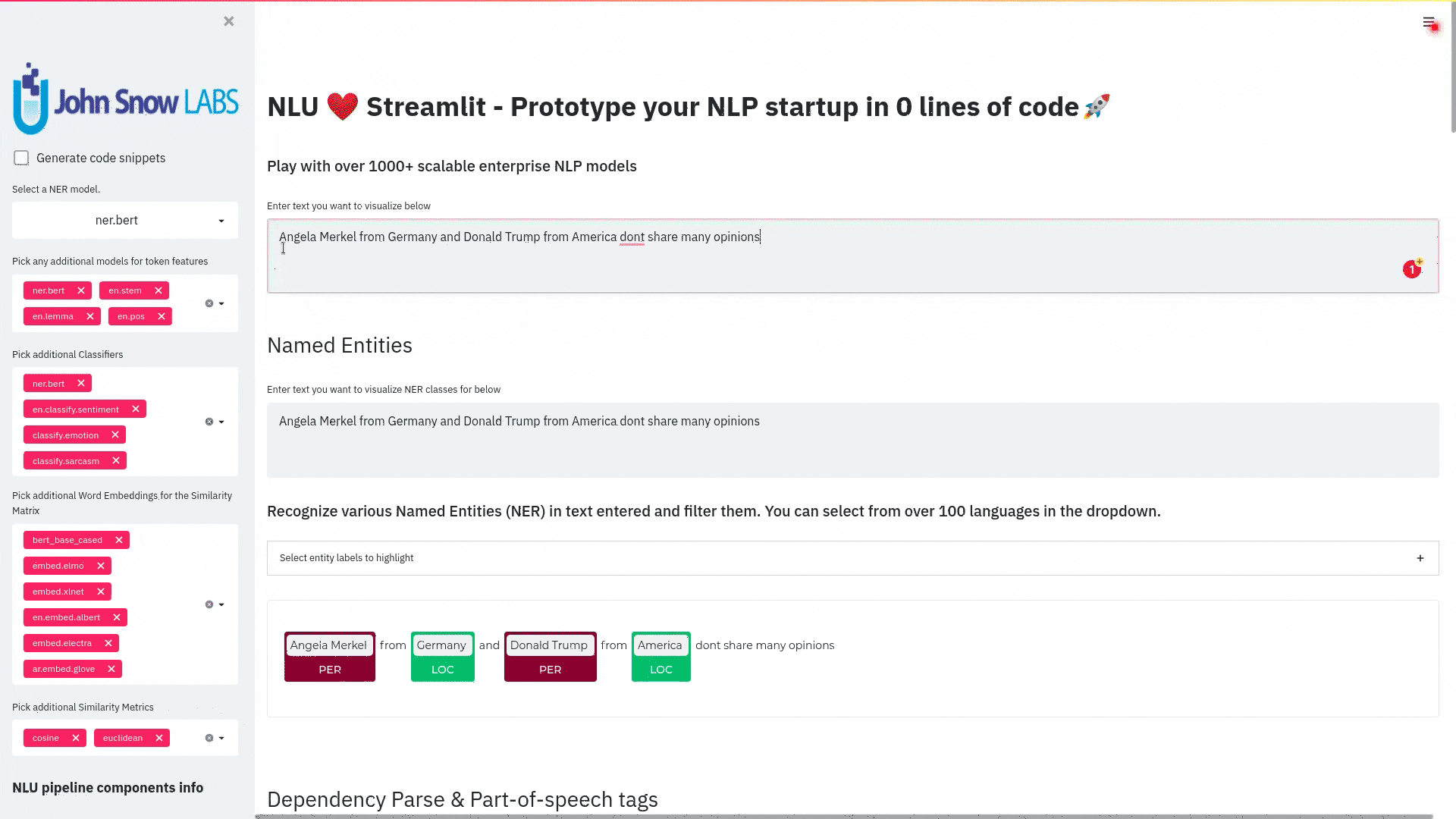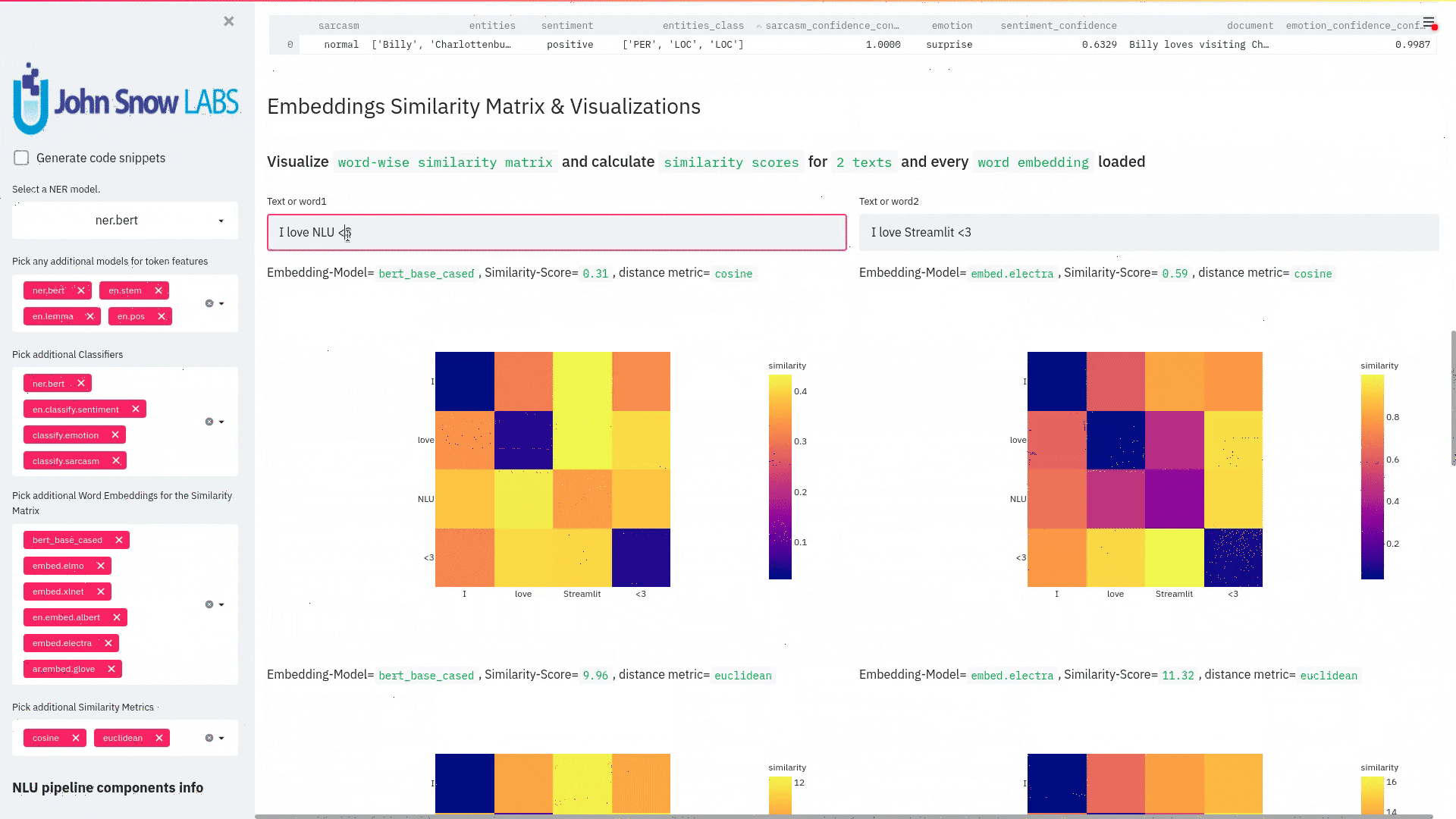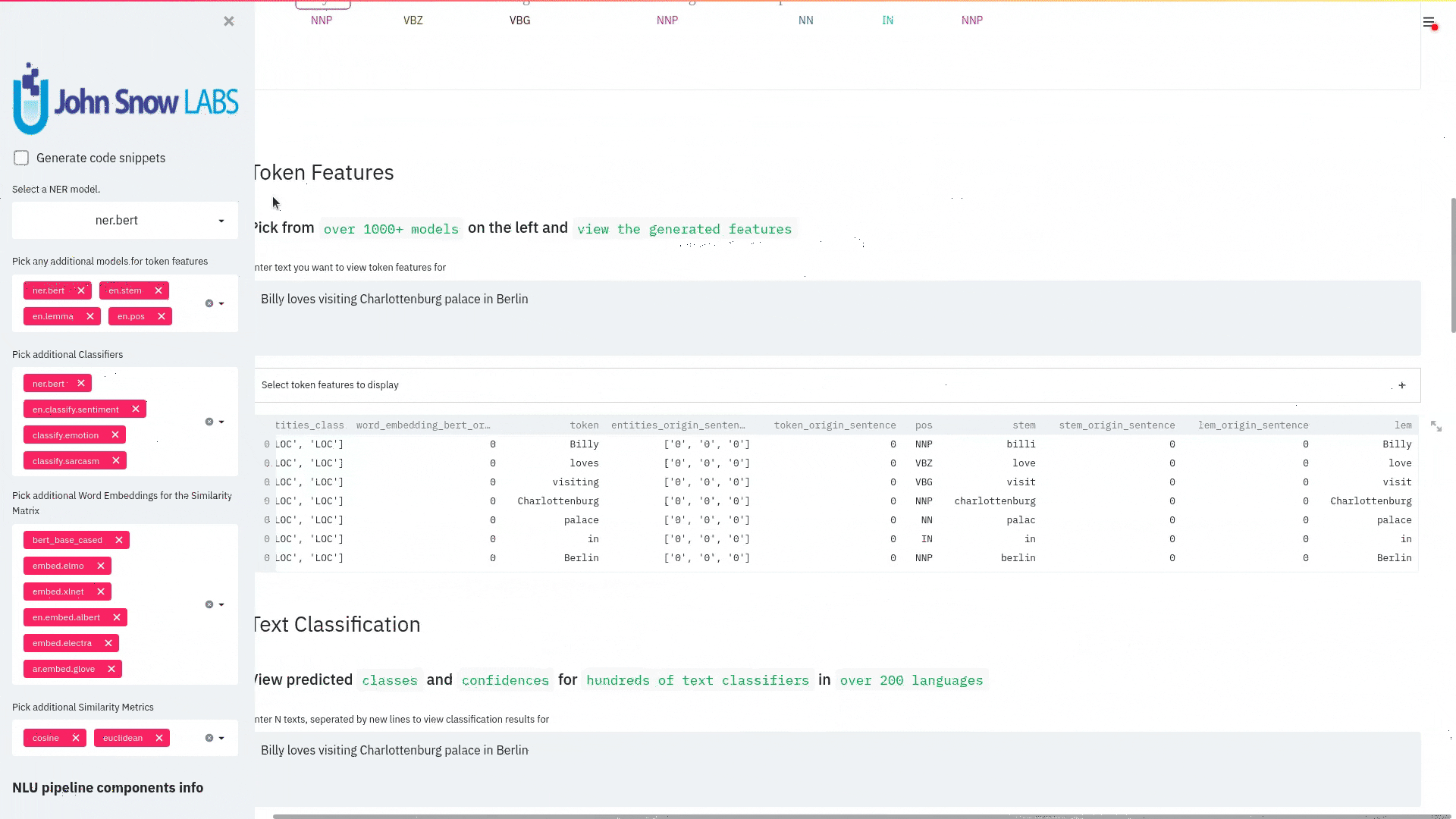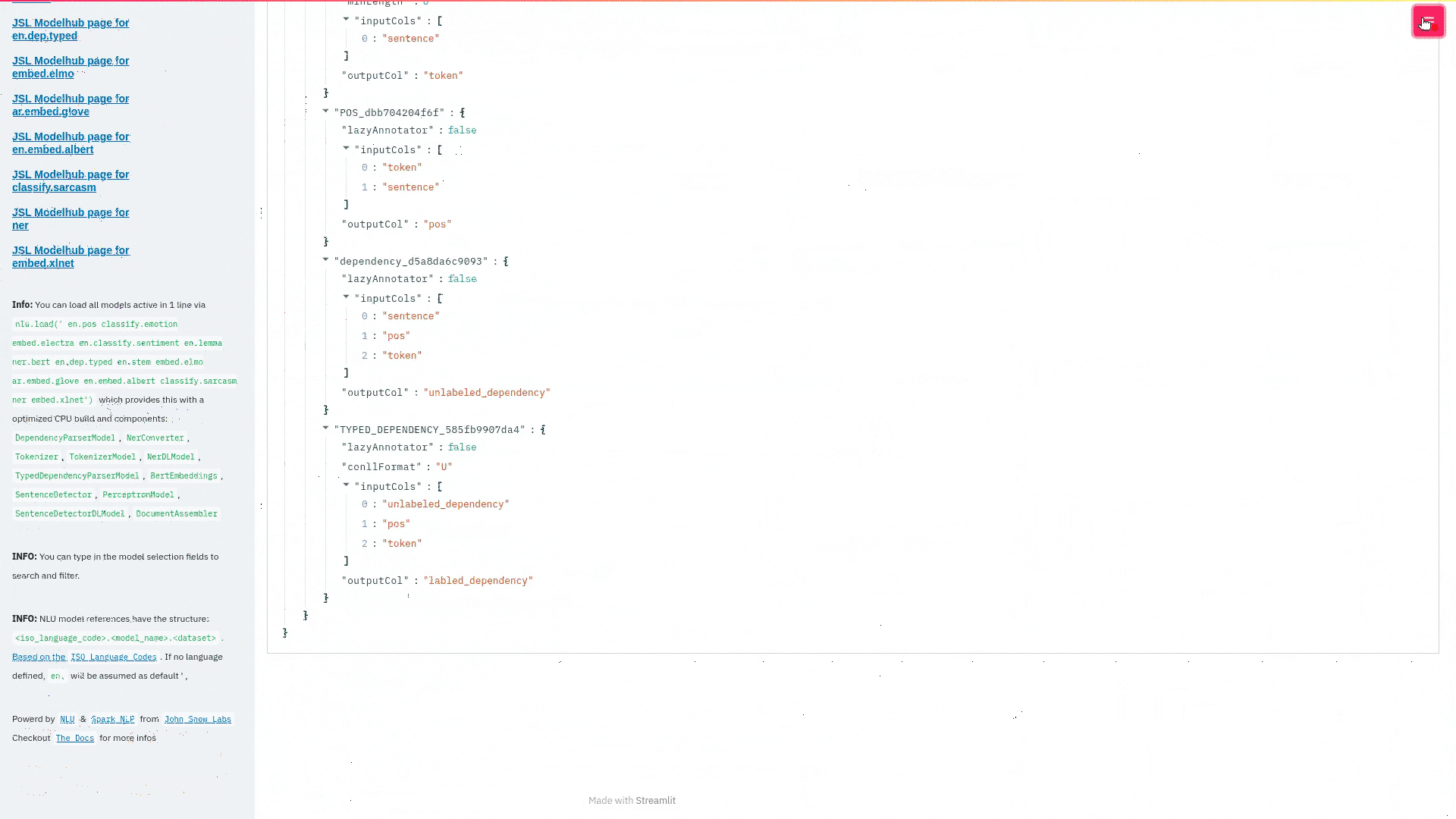
function parameters pipe.viz_streamlit
| Argument | Type | Default | Description |
|---|---|---|---|
text |
Union [str, List[str], pd.DataFrame, pd.Series] |
'NLU and Streamlit go together like peanutbutter and jelly' |
Default text for the Classification, Named Entitiy Recognizer, Token Information and Dependency Tree visualizations |
similarity_texts |
Union[List[str],Tuple[str,str]] |
('Donald Trump Likes to part', 'Angela Merkel likes to party') |
Default texts for the Text similarity visualization. Should contain exactly 2 strings which will be compared token embedding wise. For each embedding active, a token wise similarity matrix and a similarity scalar |
model_selection |
List[str] |
[] |
List of nlu references to display in the model selector, see the NLU Namespace or the John Snow Labs Modelshub or go straight to the source for more info |
title |
str |
'NLU ❤️ Streamlit - Prototype your NLP startup in 0 lines of code🚀' |
Title of the Streamlit app |
sub_title |
str |
'Play with over 1000+ scalable enterprise NLP models' |
Sub title of the Streamlit app |
visualizers |
List[str] |
( "dependency_tree", "ner", "similarity", "token_information", 'classification') |
Define which visualizations should be displayed. By default all visualizations are displayed. |
show_models_info |
bool |
True |
Show information for every model loaded in the bottom of the Streamlit app. |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
show_viz_selection |
bool |
False |
Show a selector in the sidebar which lets you configure which visualizations are displayed. |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
show_code_snippets |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
num_similarity_cols |
int |
2 |
How many columns should for the layout in Streamlit when rendering the similarity matrixes. |
function pipe.viz_streamlit_classes
Visualize the predicted classes and their confidences and additional metadata to streamlit. Aplicable with any of the 100+ classifiers
nlu.load('sentiment').viz_streamlit_classes(['I love NLU and Streamlit!','I love buggy software', 'Sign up now get a chance to win 1000$ !', 'I am afraid of Snakes','Unicorns have been sighted on Mars!','Where is the next bus stop?'])
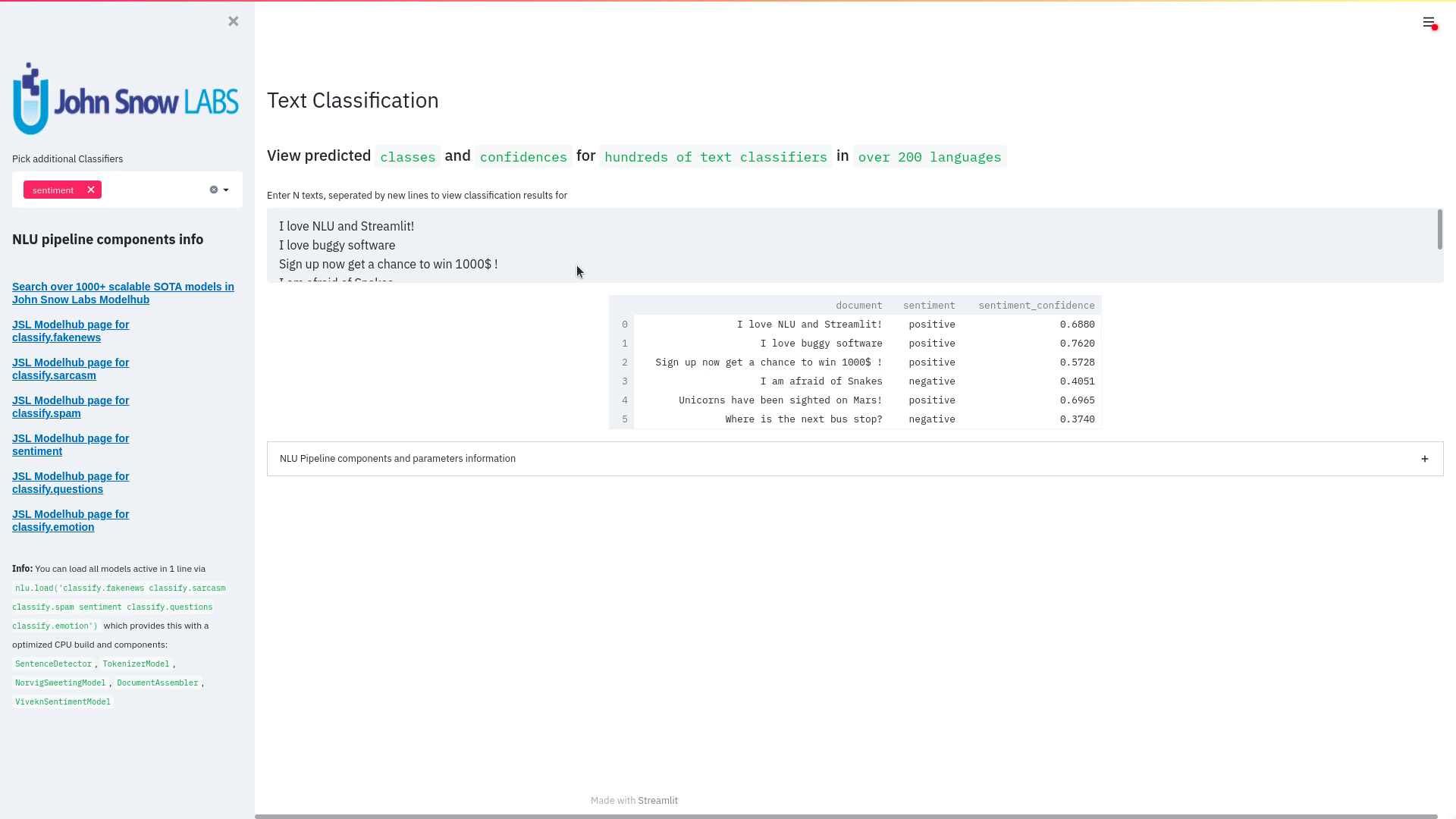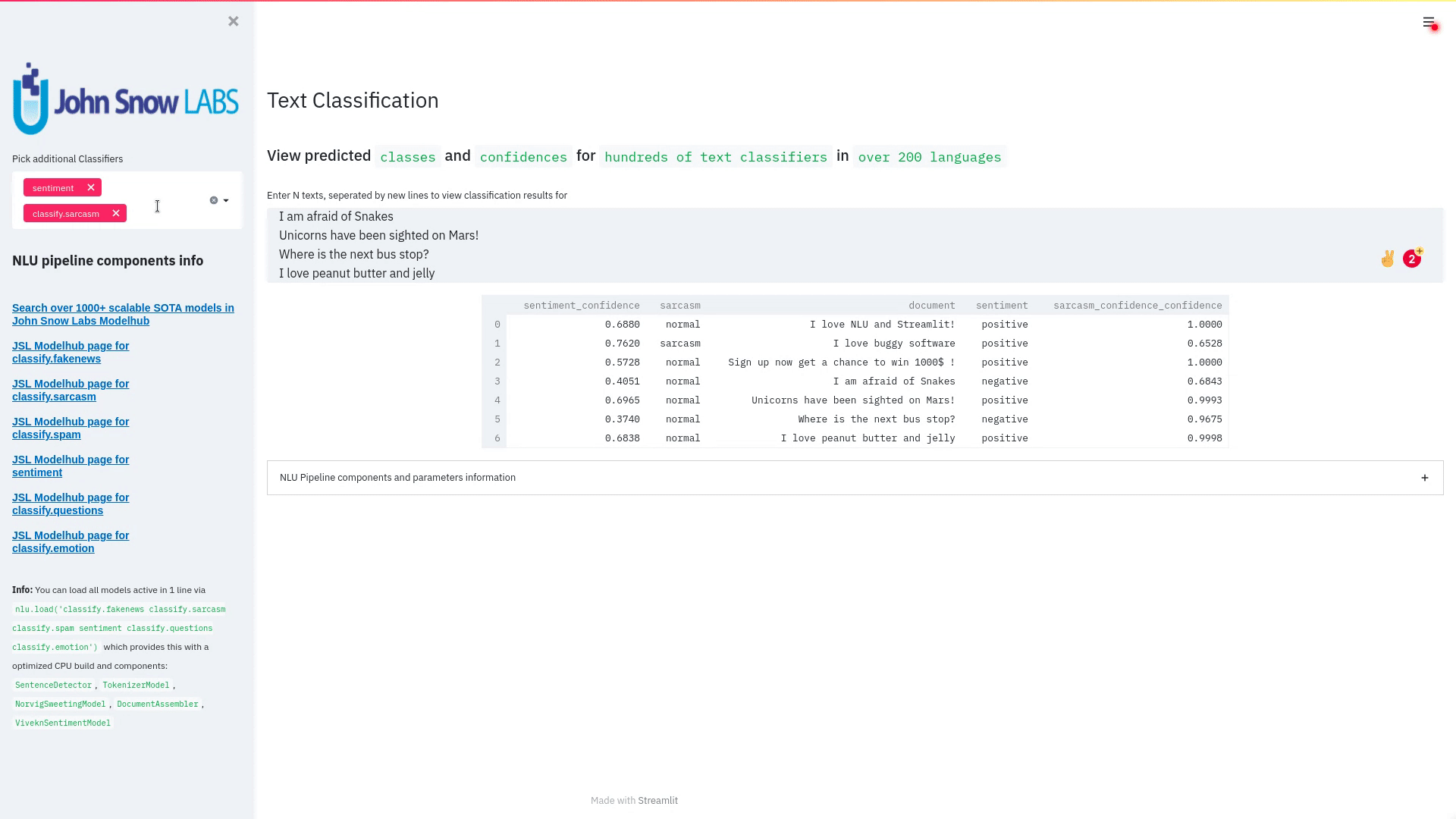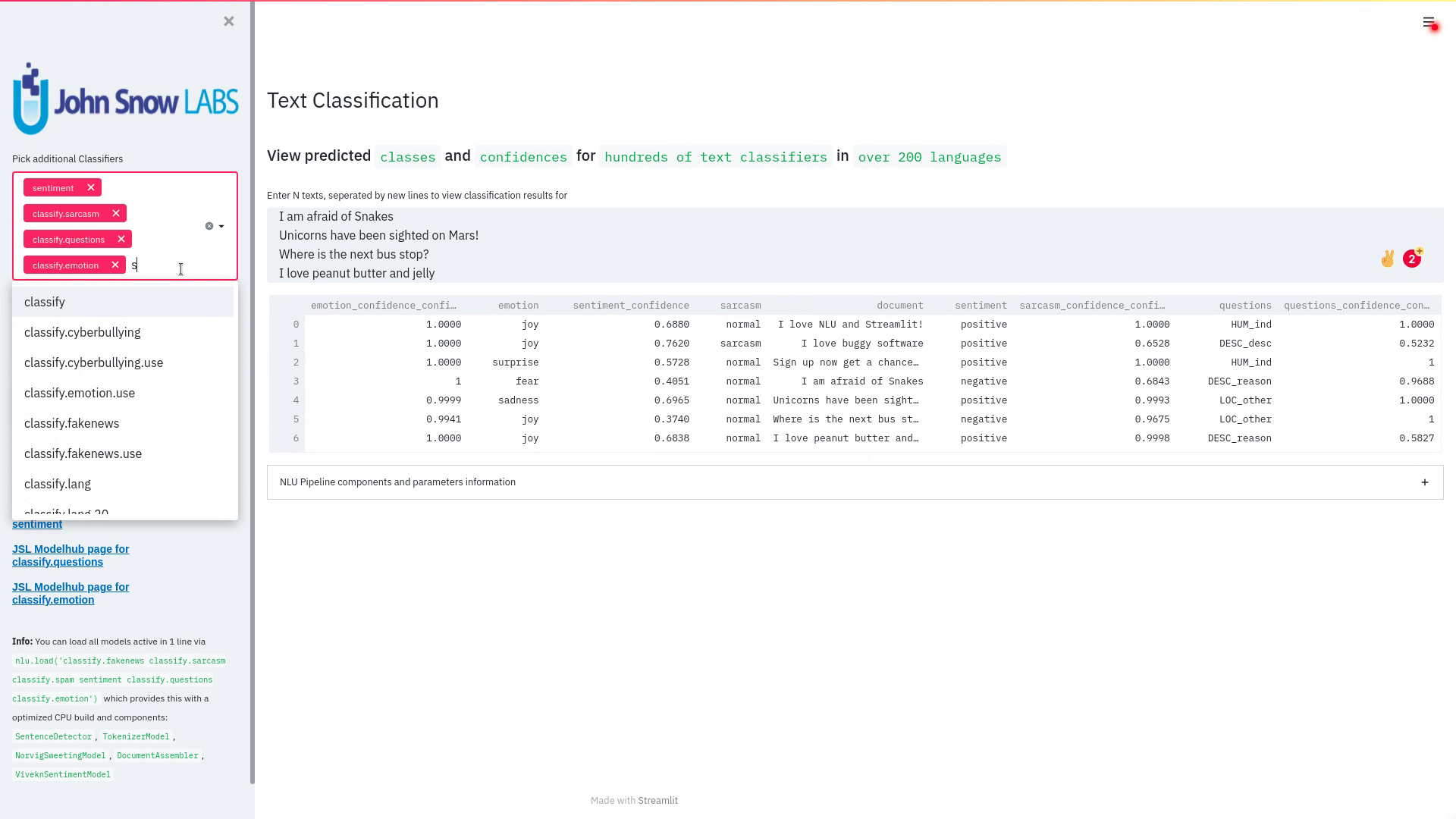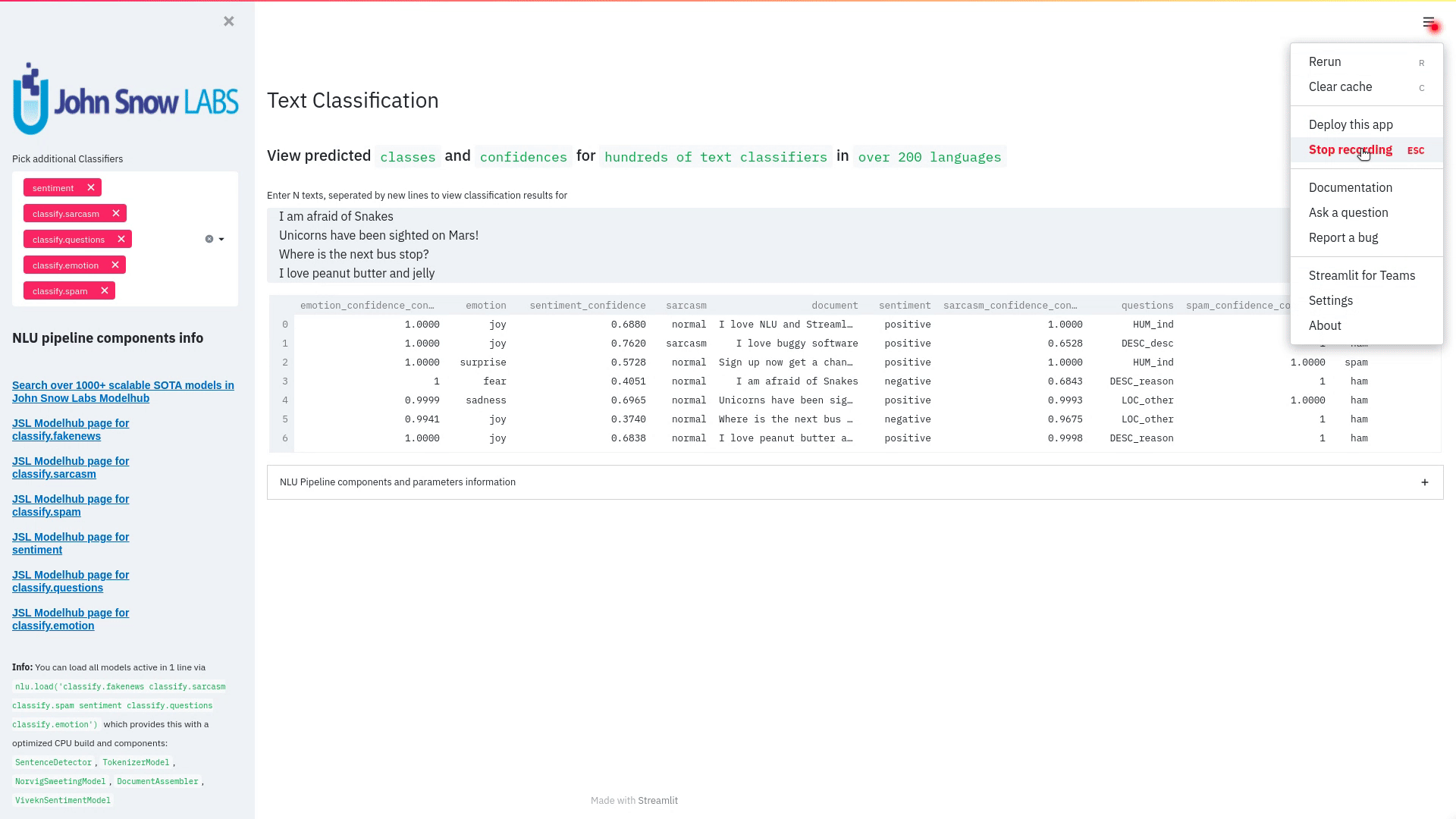
function parameters pipe.viz_streamlit_classes
| Argument | Type | Default | Description |
|---|---|---|---|
text |
Union[str,list,pd.DataFrame, pd.Series, pyspark.sql.DataFrame ] |
'I love NLU and Streamlit and sunny days!' |
Text to predict classes for. Will predict on each input of the iteratable or dataframe if type is not str. |
output_level |
Optional[str] |
document |
Outputlevel of NLU pipeline, see pipe.predict() docsmore info |
include_text_col |
bool |
True |
Whether to include a e text column in the output table or just the prediction data |
title |
Optional[str] |
Text Classification |
Title of the Streamlit building block that will be visualized to screen |
metadata |
bool |
False |
whether to output addition metadata or not, see pipe.predict(meta=true) docs for more info |
positions |
bool |
False |
whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
function pipe.viz_streamlit_ner
Visualize the predicted classes and their confidences and additional metadata to Streamlit.
Aplicable with any of the 250+ NER models.
You can filter which NER tags to highlight via the dropdown in the main window.
Basic usage
nlu.load('ner').viz_streamlit_ner('Donald Trump from America and Angela Merkel from Germany dont share many views')
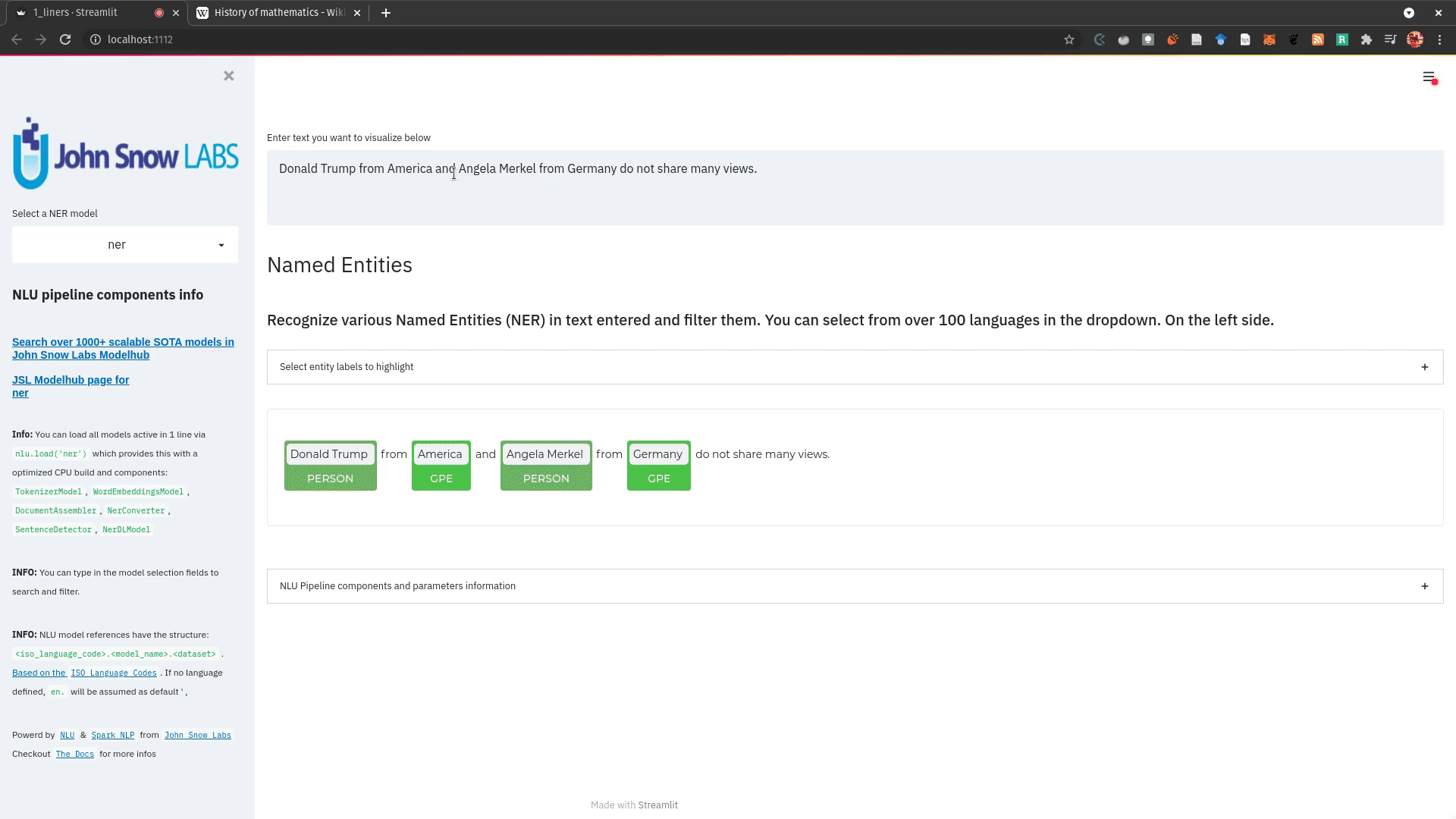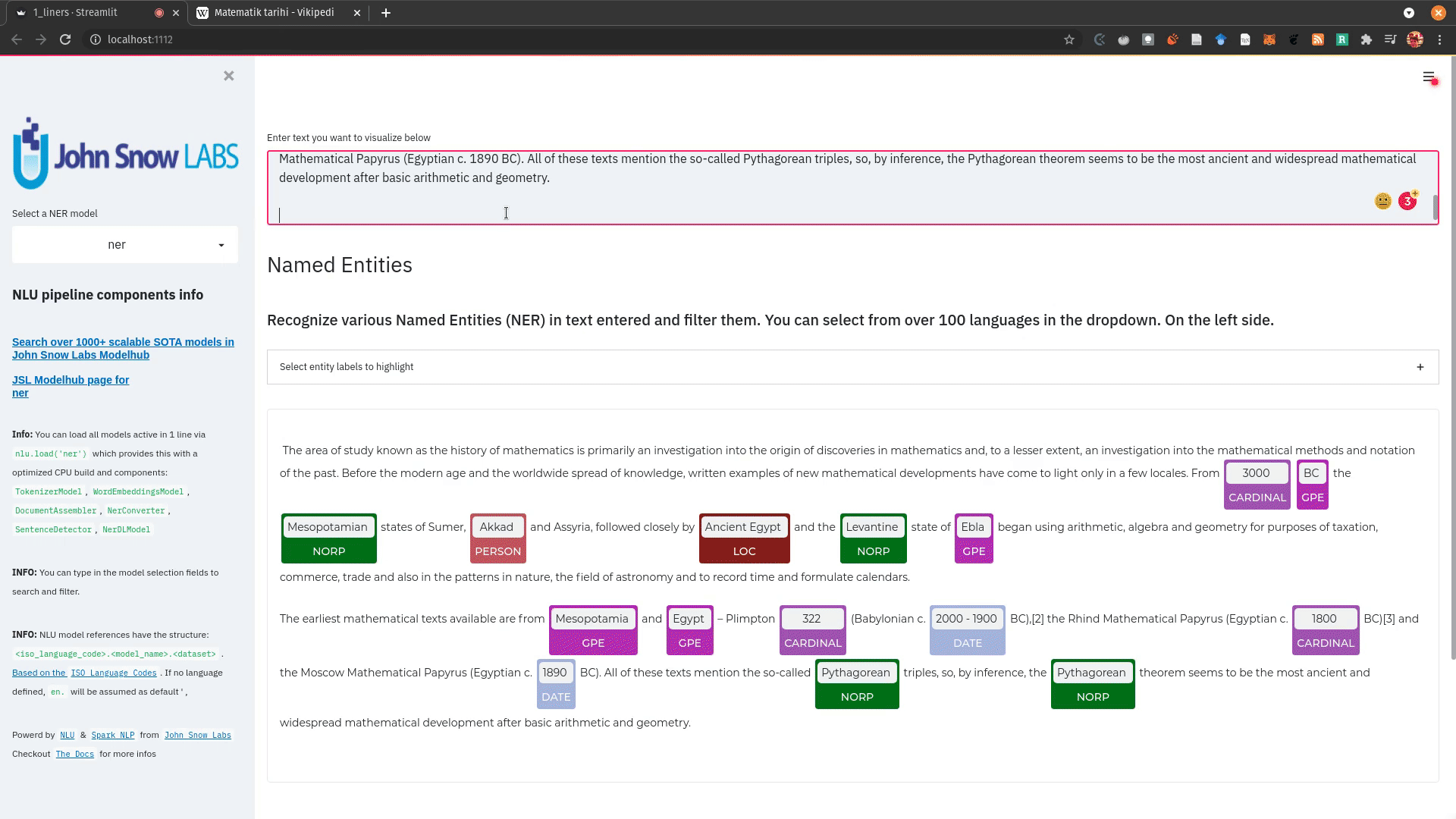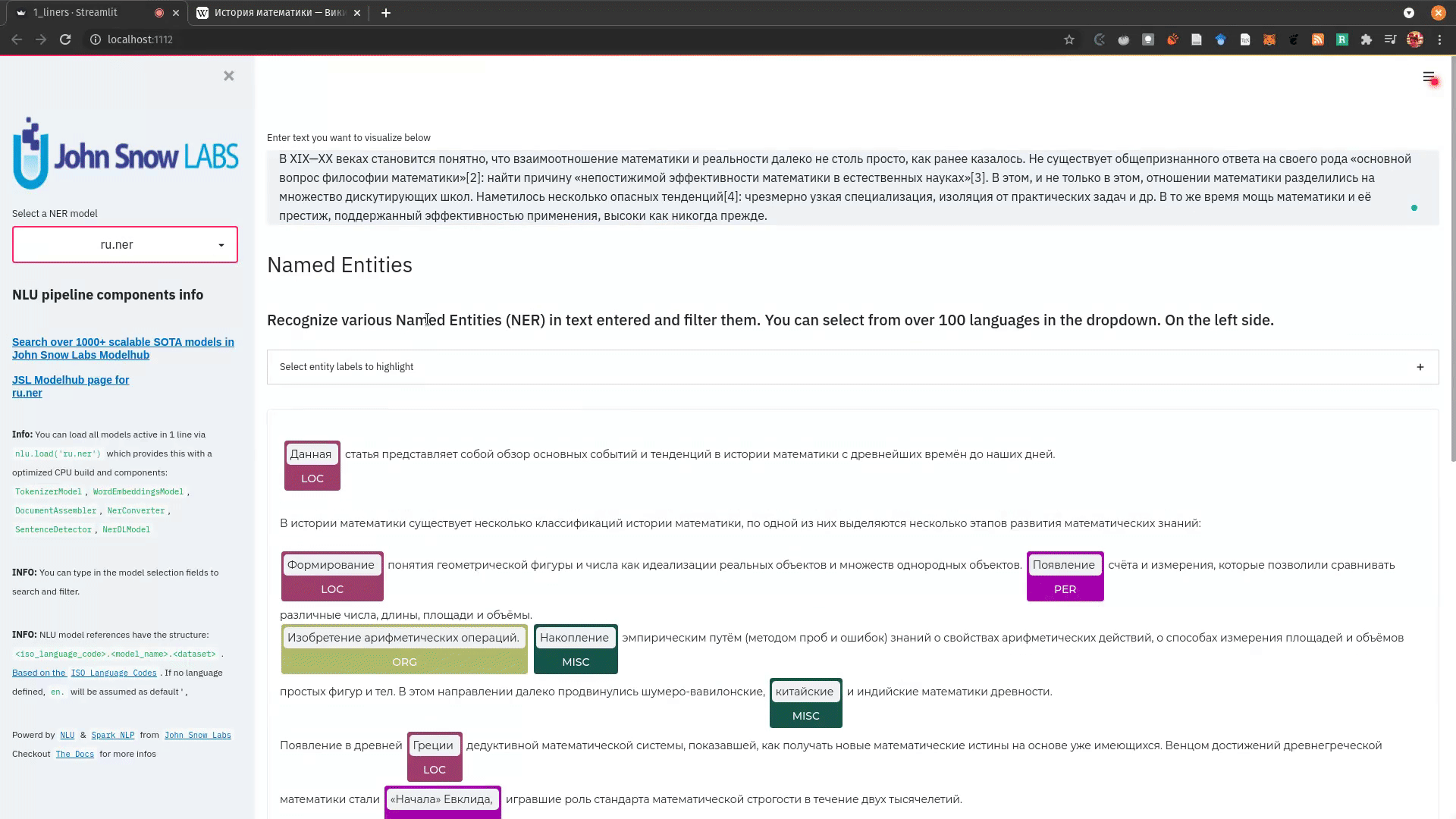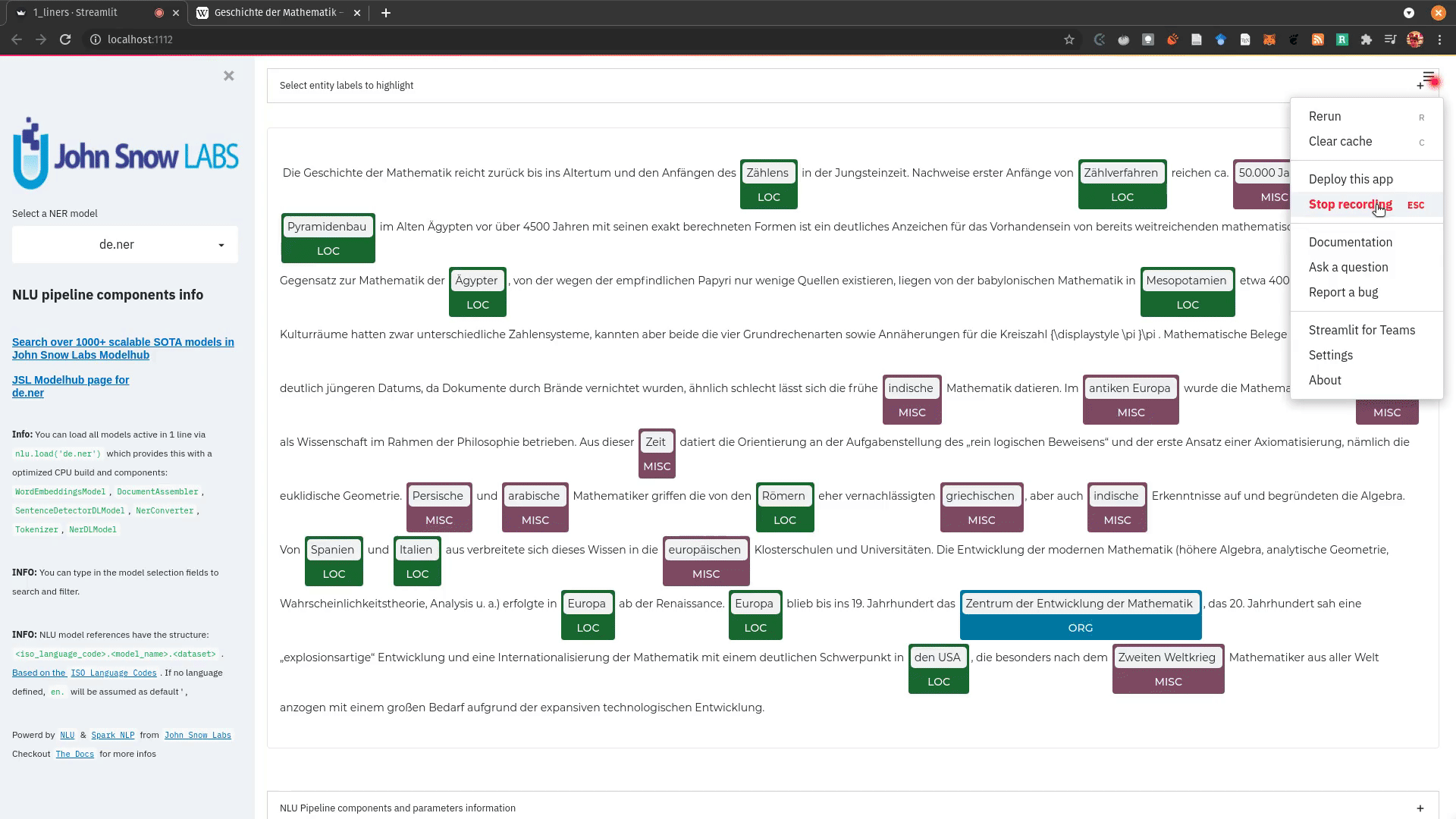
Example for coloring
# Color all entities of class GPE black
nlu.load('ner').viz_streamlit_ner('Donald Trump from America and Angela Merkel from Germany dont share many views',colors={'PERSON':'#6e992e', 'GPE':'#000000'})
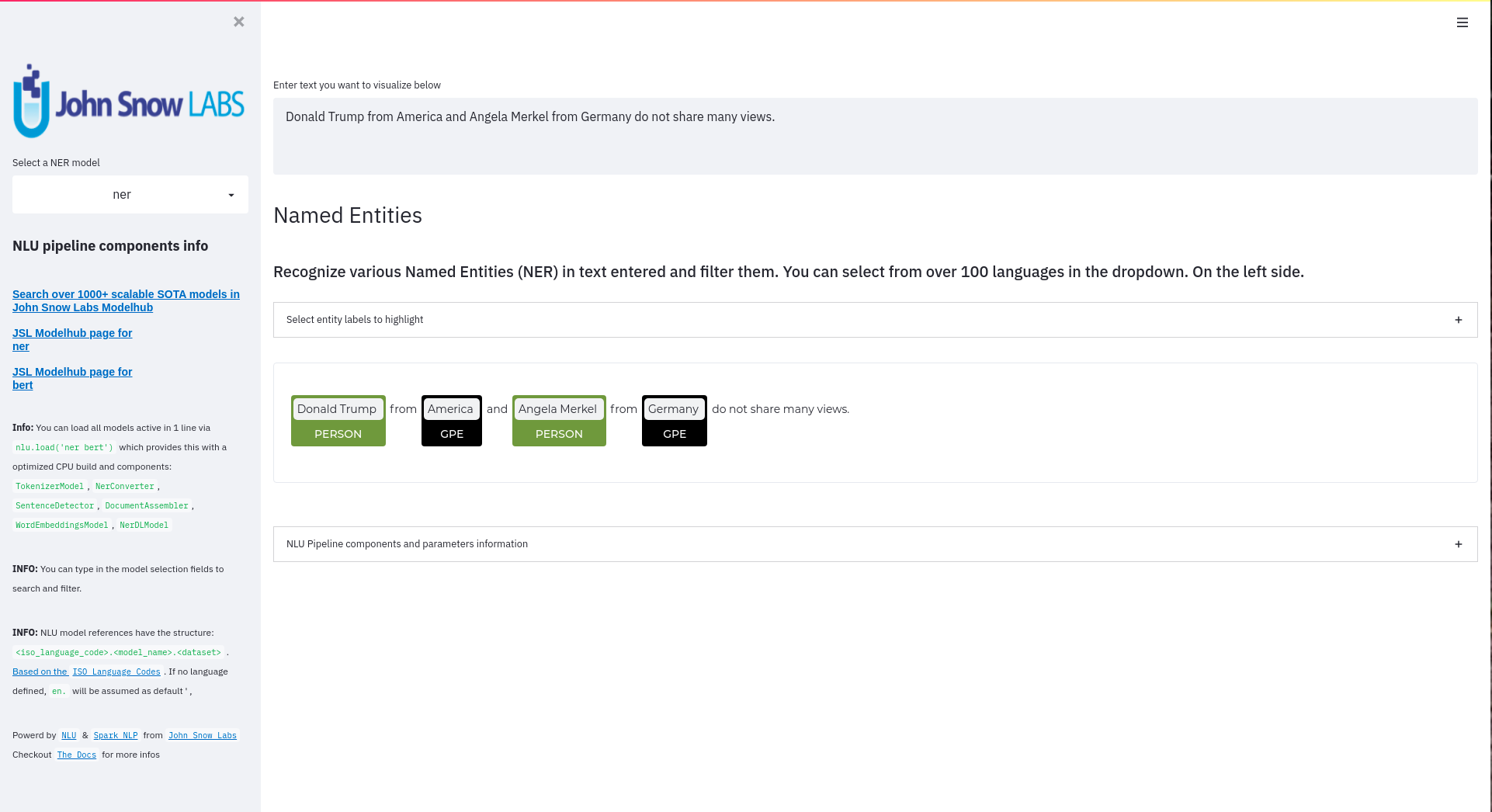
function parameters pipe.viz_streamlit_ner
| Argument | Type | Default | Description |
|---|---|---|---|
text |
str |
'Donald Trump from America and Anegela Merkel from Germany do not share many views' |
Text to predict classes for. |
ner_tags |
Optional[List[str]] |
None |
Tags to display. By default all tags will be displayed |
show_label_select |
bool |
True |
Whether to include the label selector |
show_table |
bool |
True |
Whether show to predicted pandas table or not |
title |
Optional[str] |
'Named Entities' |
Title of the Streamlit building block that will be visualized to screen |
sub_title |
Optional[str] |
'"Recognize various Named Entities (NER) in text entered and filter them. You can select from over 100 languages in the dropdown. On the left side.",' |
Sub-title of the Streamlit building block that will be visualized to screen |
colors |
Dict[str,str] |
{} |
Dict with KEY=ENTITY_LABEL and VALUE=COLOR_AS_HEX_CODE,which will change color of highlighted entities.See custom color labels docs for more info. |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
show_text_input |
bool |
True |
Show text input field to input text in |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
function pipe.viz_streamlit_dep_tree
Visualize a typed dependency tree, the relations between tokens and part of speech tags predicted. Aplicable with any of the 100+ Part of Speech(POS) models and dep tree model
nlu.load('dep.typed').viz_streamlit_dep_tree('POS tags define a grammatical label for each token and the Dependency Tree classifies Relations between the tokens')
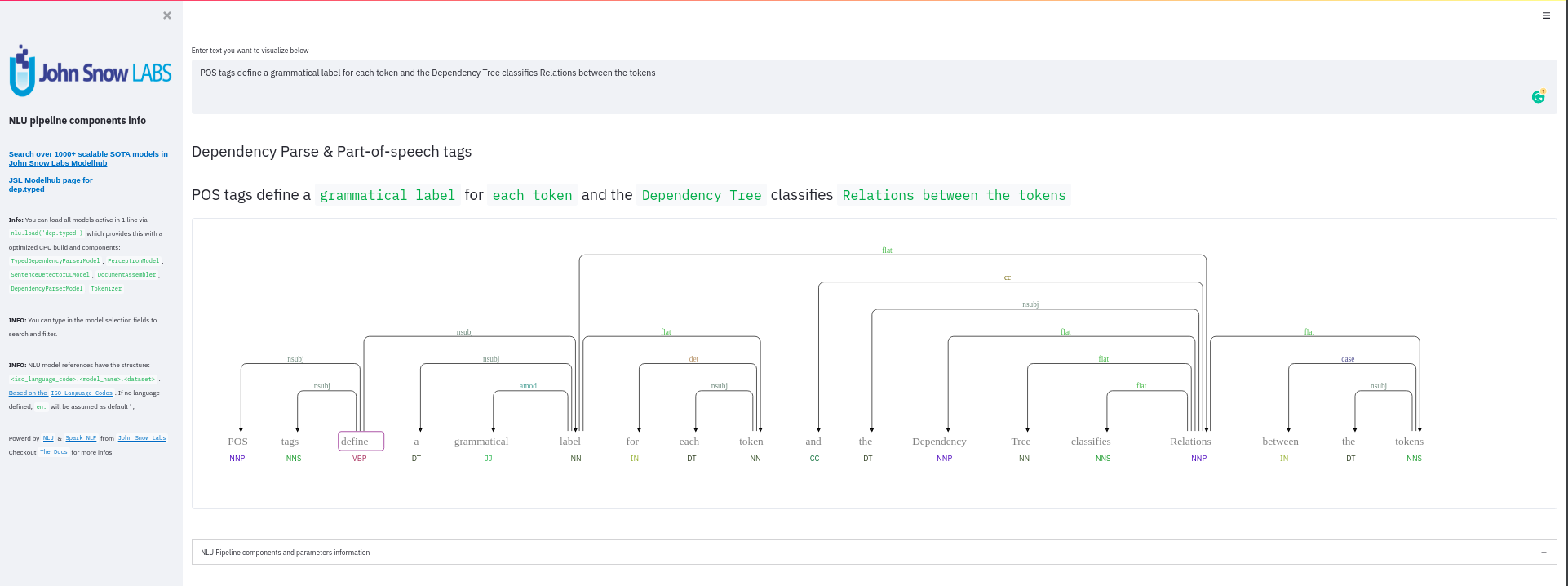
function parameters pipe.viz_streamlit_dep_tree
| Argument | Type | Default | Description |
|---|---|---|---|
text |
str |
'Billy likes to swim' |
Text to predict classes for. |
title |
Optional[str] |
'Dependency Parse Tree & Part-of-speech tags' |
Title of the Streamlit building block that will be visualized to screen |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
function pipe.viz_streamlit_token
Visualize predicted token and text features for every model loaded. You can use this with any of the 1000+ models and select them from the left dropdown.
nlu.load('stemm pos spell').viz_streamlit_token('I liek pentut buttr and jelly !')
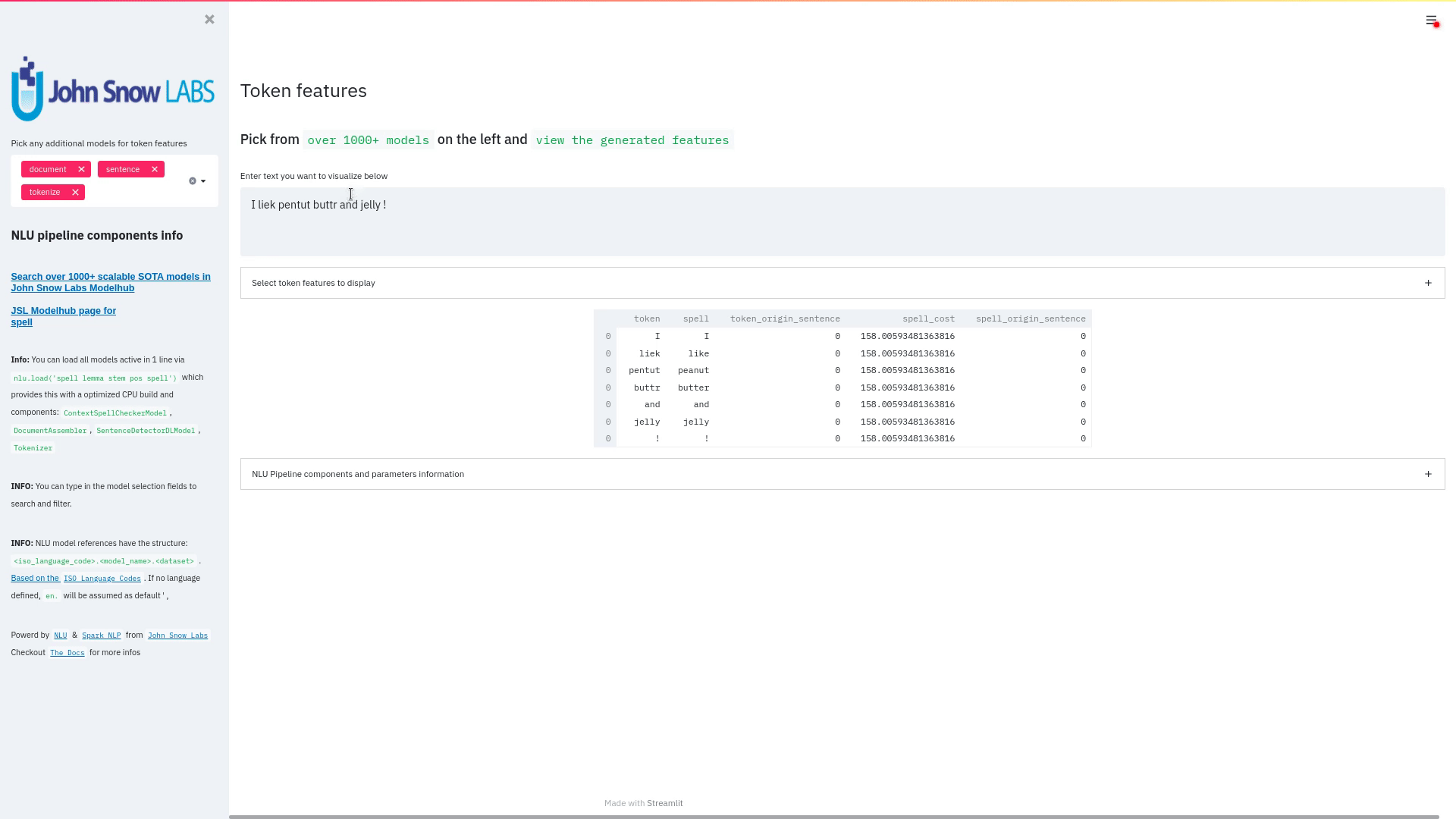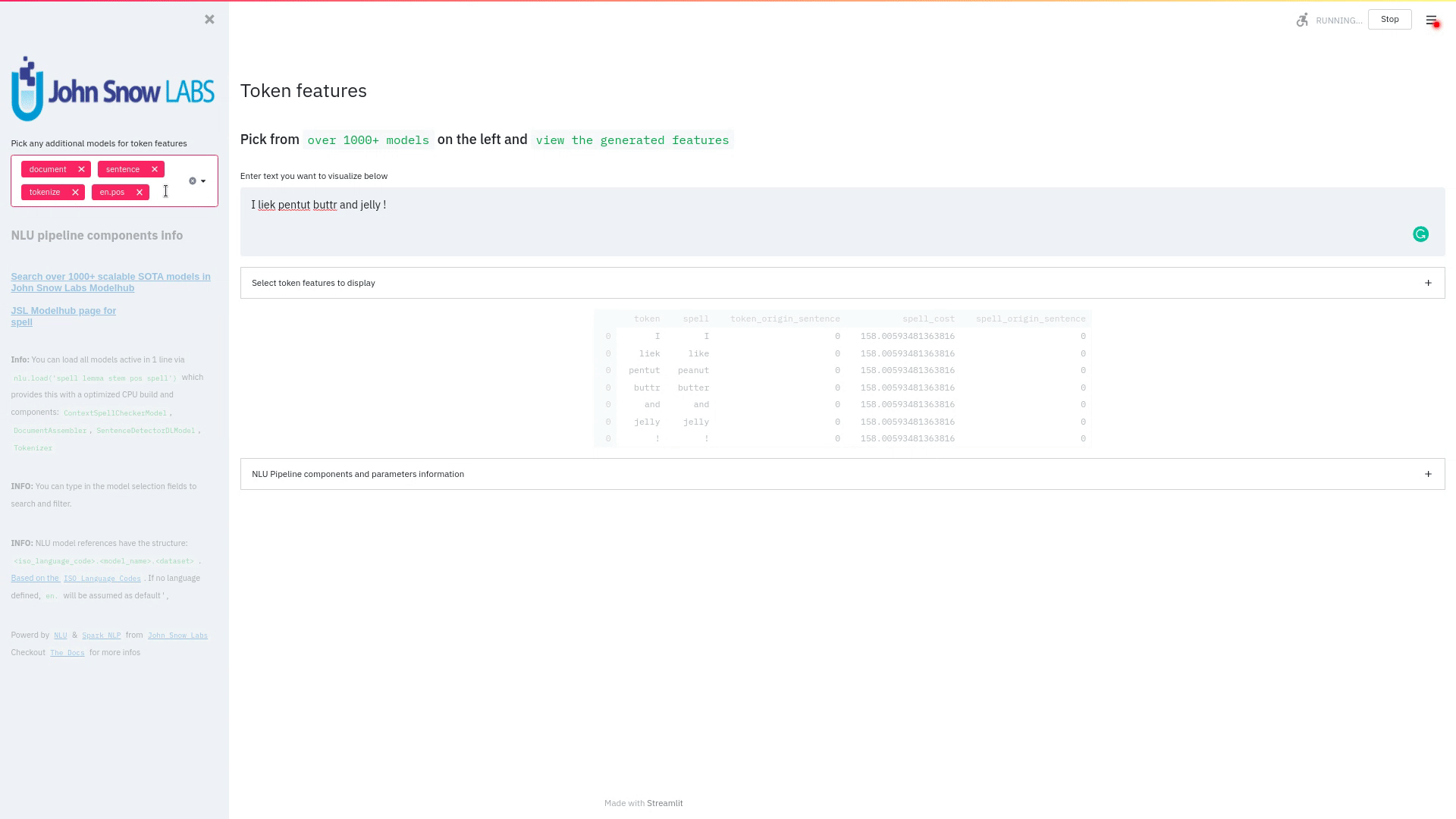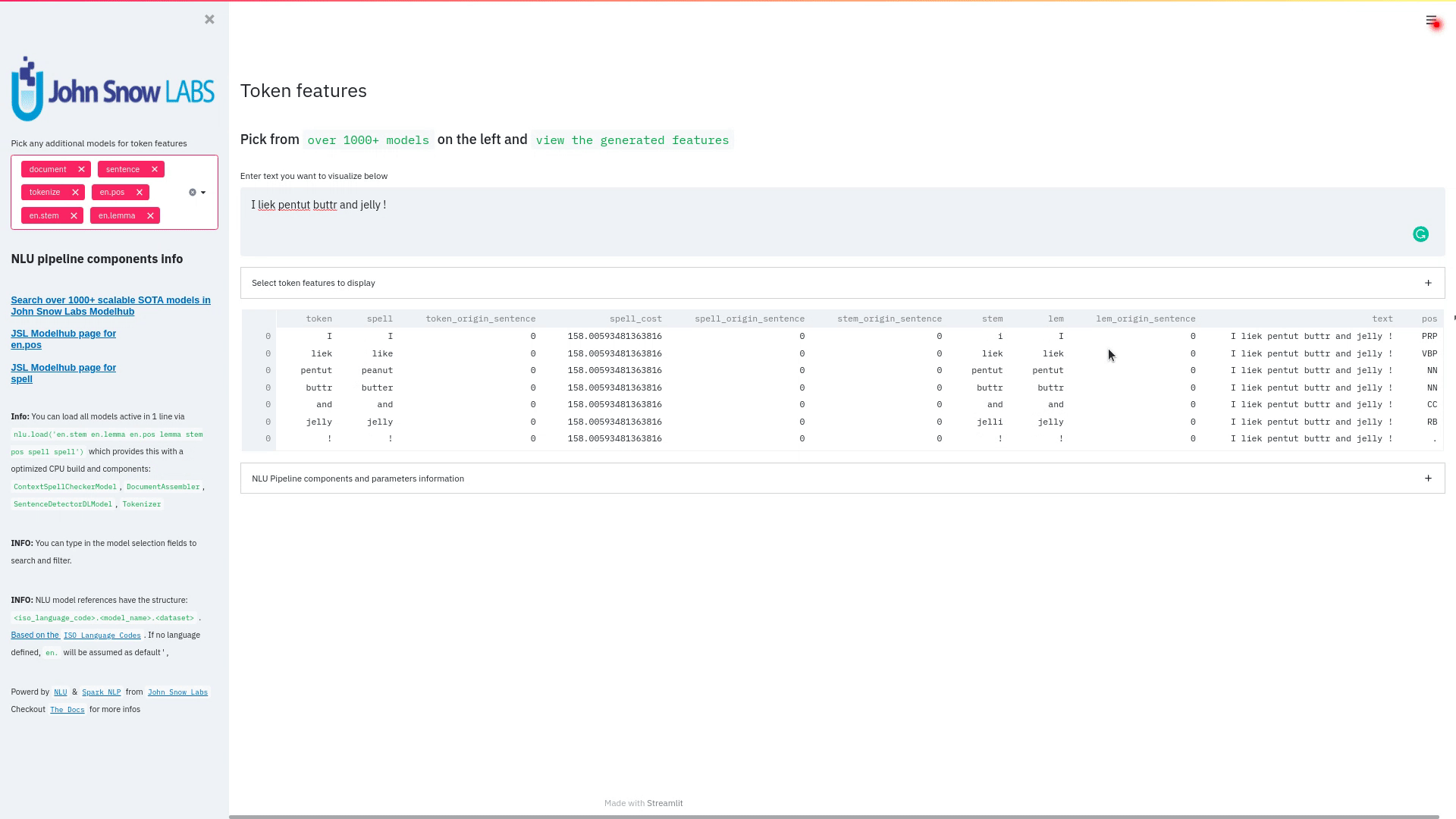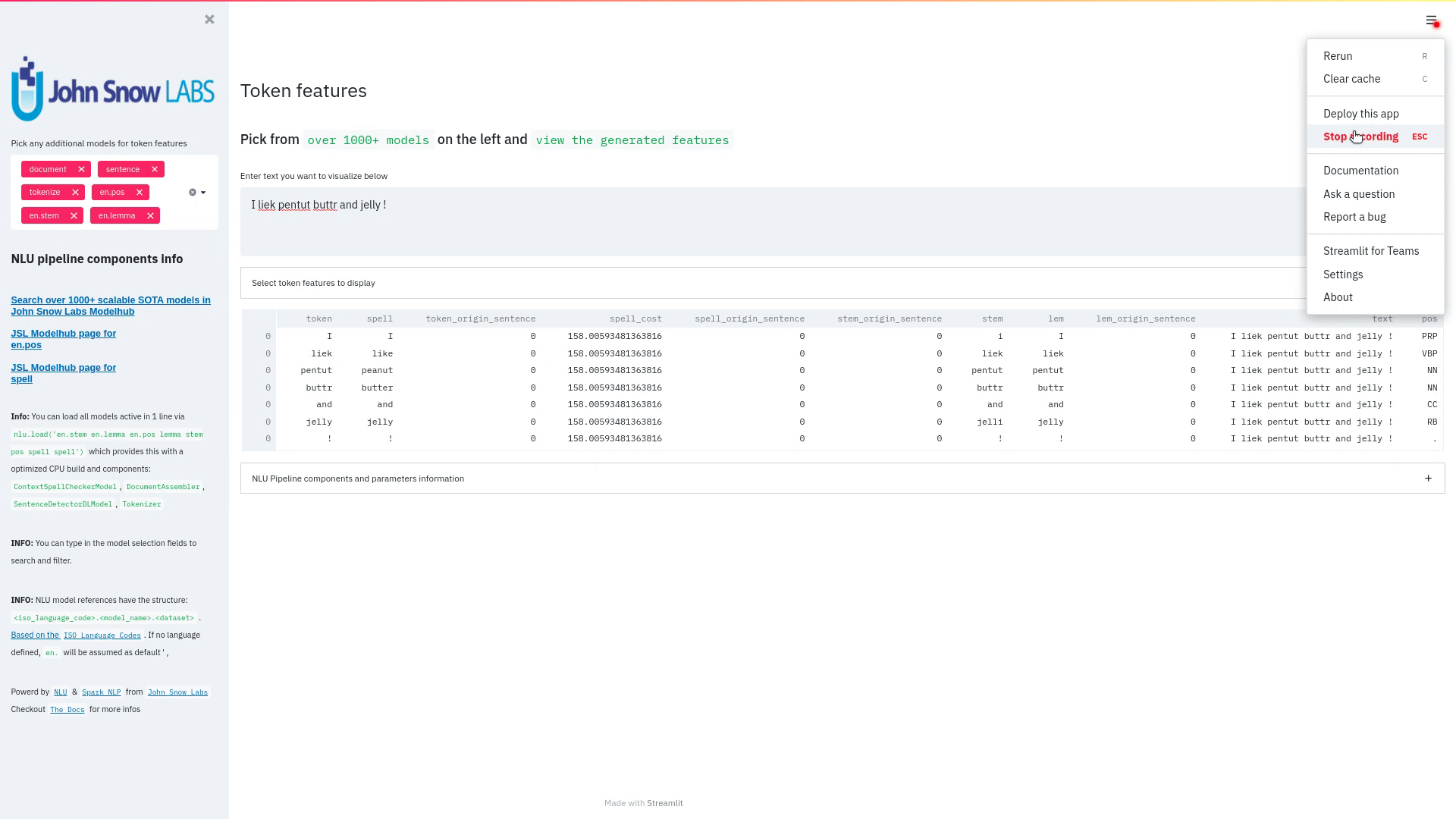
function parameters pipe.viz_streamlit_token
| Argument | Type | Default | Description |
|---|---|---|---|
text |
str |
'NLU and Streamlit are great!' |
Text to predict token information for. |
title |
Optional[str] |
'Named Entities' |
Title of the Streamlit building block that will be visualized to screen |
show_feature_select |
bool |
True |
Whether to include the token feature selector |
features |
Optional[List[str]] |
None |
Features to to display. By default all Features will be displayed |
metadata |
bool |
False |
Whether to output addition metadata or not, see pipe.predict(meta=true) docs for more info |
output_level |
Optional[str] |
'token' |
Outputlevel of NLU pipeline, see pipe.predict() docsmore info |
positions |
bool |
False |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
function pipe.viz_streamlit_similarity
- Displays a
similarity matrix, wherex-axisis every token in the first text andy-axisis every token in the second text. - Index
i,jin the matrix describes the similarity oftoken-itotoken-jbased on the loaded embeddings and distance metrics, based on Sklearns Pariwise Metrics.. See this article for more elaboration on similarities - Displays a dropdown selectors from which various similarity metrics and over 100 embeddings can be selected.
-There will be one similarity matrix per
metricandembeddingpair selected.num_plots = num_metric*num_embeddingsAlso displays embedding vector information. Applicable with any of the 100+ Word Embedding models
nlu.load('bert').viz_streamlit_word_similarity(['I love love loooove NLU! <3','I also love love looove Streamlit! <3'])
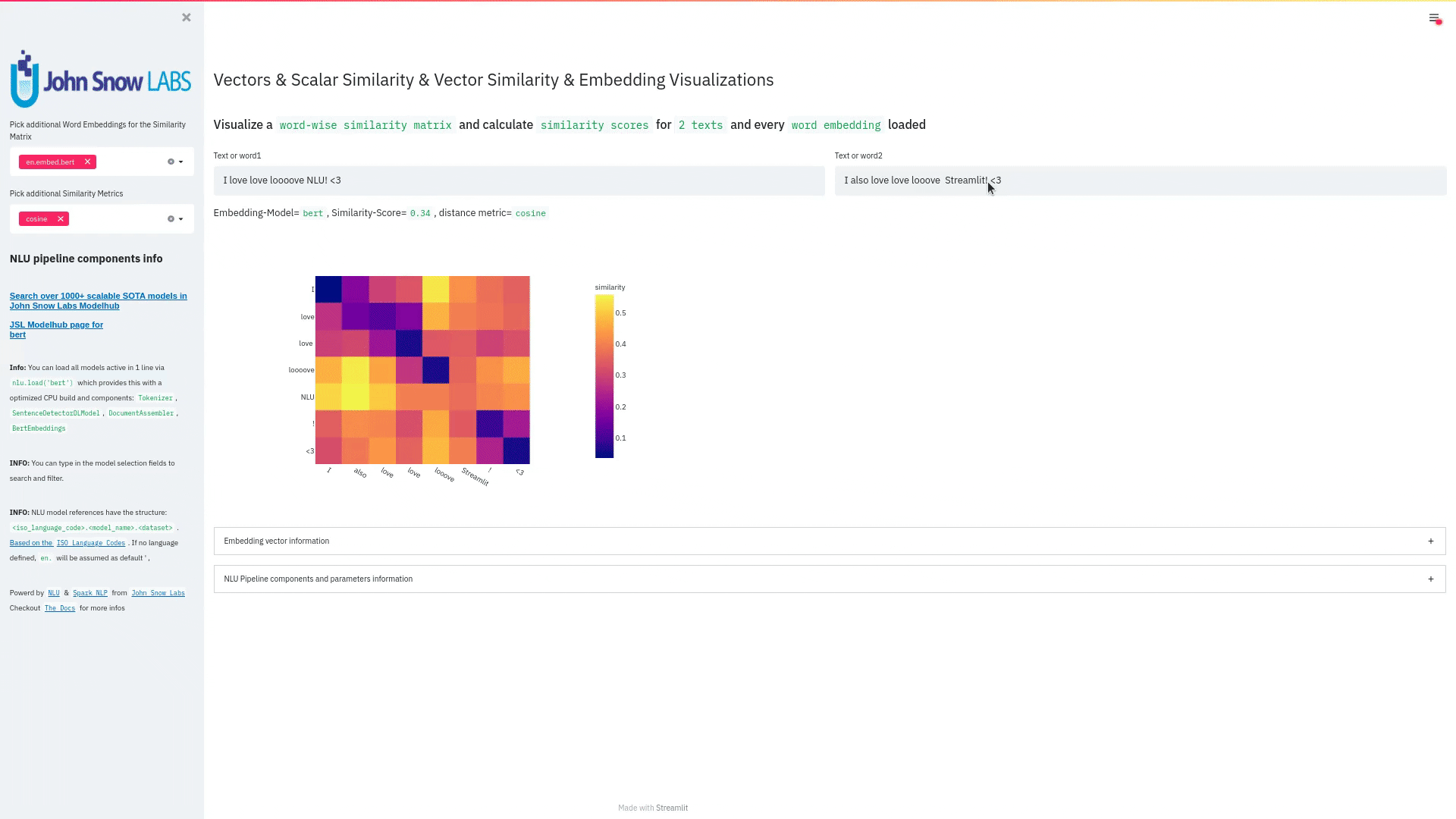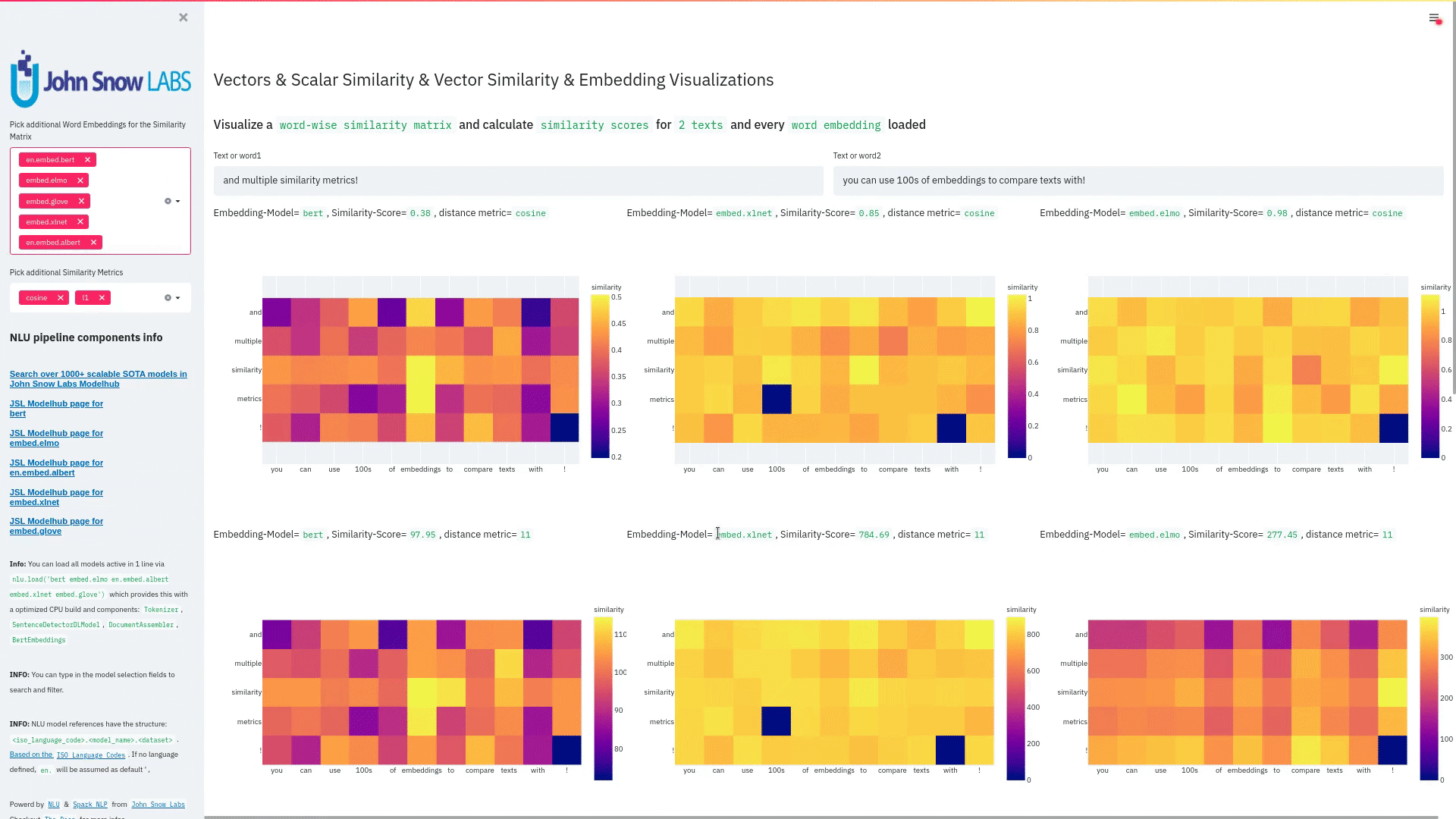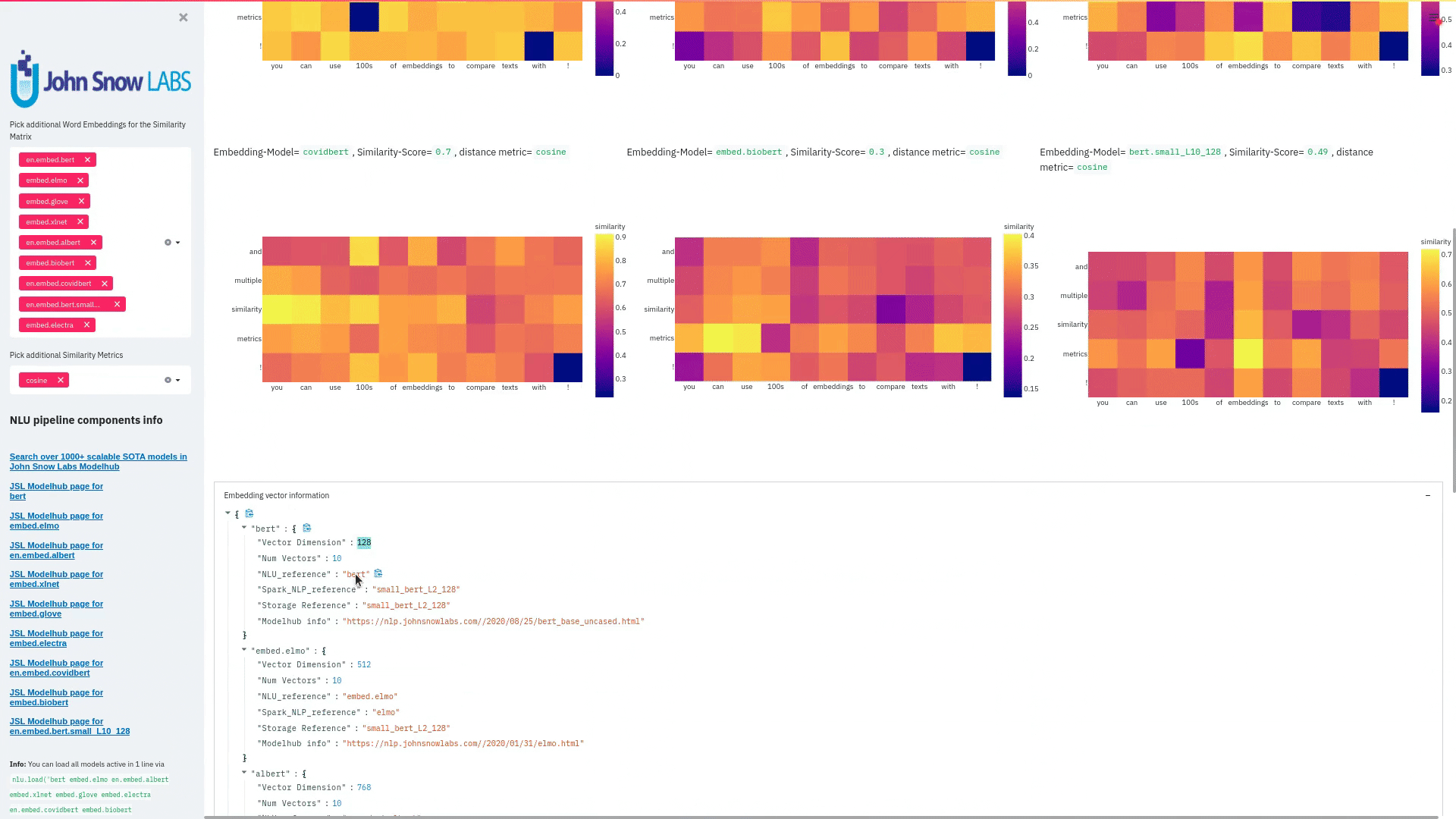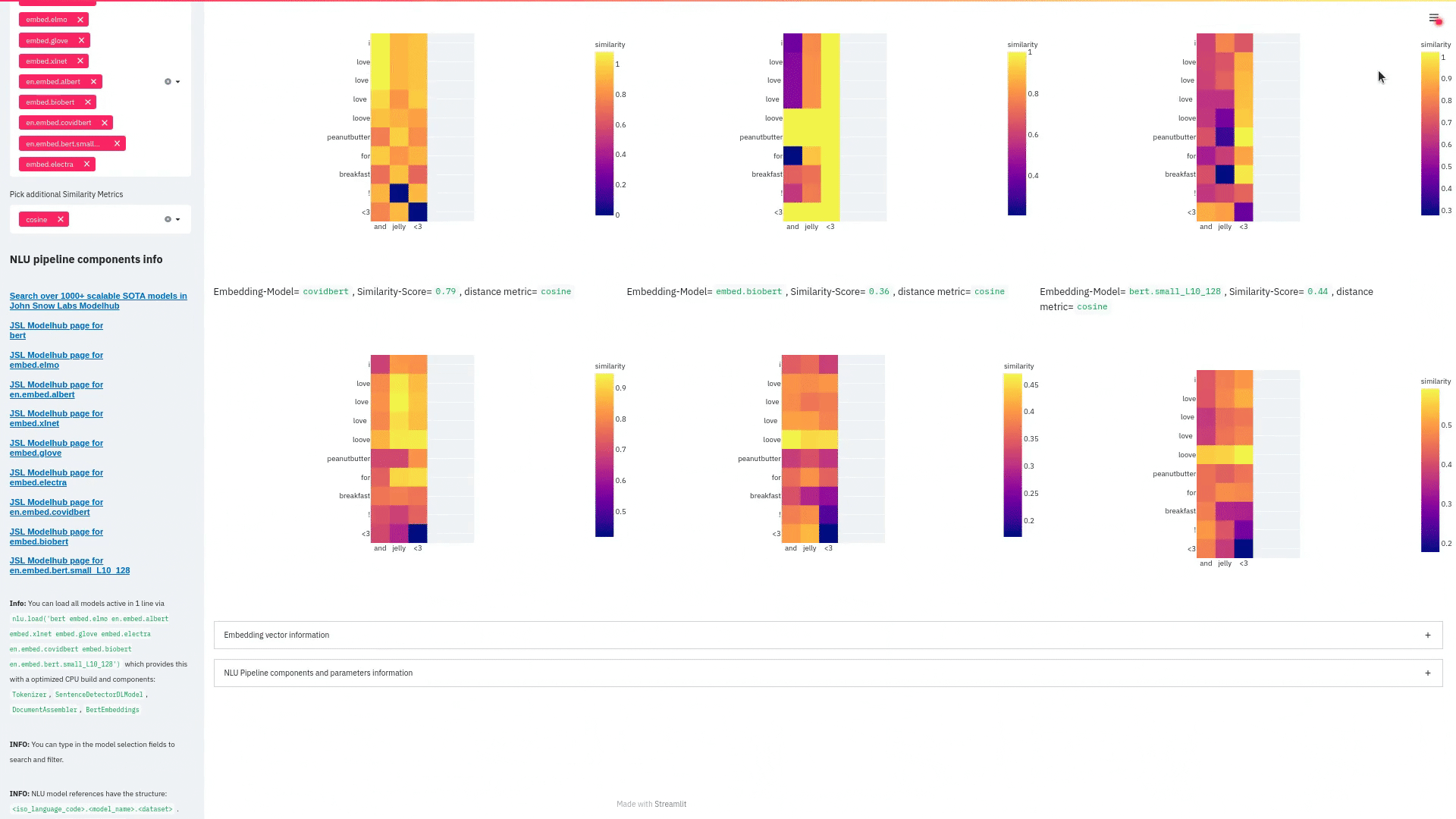
function parameters pipe.viz_streamlit_similarity
| Argument | Type | Default | Description |
|---|---|---|---|
texts |
str |
'Donald Trump from America and Anegela Merkel from Germany do not share many views.' |
Text to predict token information for. |
title |
Optional[str] |
'Named Entities' |
Title of the Streamlit building block that will be visualized to screen |
similarity_matrix |
bool |
None |
Whether to display similarity matrix or not |
show_algo_select |
bool |
True |
Whether to show dist algo select or not |
show_table |
bool |
True |
Whether show to predicted pandas table or not |
threshold |
float |
0.5 |
Threshold for displaying result red on screen |
set_wide_layout_CSS |
bool |
True |
Whether to inject custom CSS or not. |
key |
str |
"NLU_streamlit" |
Key for the Streamlit elements drawn |
generate_code_sample |
bool |
False |
Display Python code snippets above visualizations that can be used to re-create the visualization |
show_model_select |
bool |
True |
Show a model selection dropdowns that makes any of the 1000+ models avaiable in 1 click |
model_select_position |
str |
'side' |
Whether to output the positions of predictions or not, see pipe.predict(positions=true) for more info |
write_raw_pandas |
bool |
False |
Write the raw pandas similarity df to streamlit |
display_embed_information |
bool |
True |
Show additional embedding information like dimension, nlu_reference, spark_nlp_reference, sotrage_reference, modelhub link and more. |
dist_metrics |
List[str] |
['cosine'] |
Which distance metrics to apply. If multiple are selected, there will be multiple plots for each embedding and metric. num_plots = num_metric*num_embeddings. Can use multiple at the same time, any of of cityblock,cosine,euclidean,l2,l1,manhattan,nan_euclidean. Provided via Sklearn metrics.pairwise package |
num_cols |
int |
2 |
How many columns should for the layout in streamlit when rendering the similarity matrixes. |
display_scalar_similarities |
bool |
False |
Display scalar simmilarities in an additional field. |
display_similarity_summary |
bool |
False |
Display summary of all similarities for all embeddings and metrics. |
show_logo |
bool |
True |
Show logo |
display_infos |
bool |
False |
Display additonal information about ISO codes and the NLU namespace structure. |
In addition have added some new features to our T5 Transformer annotator to help with longer and more accurate text generation, trained some new multi-lingual models and pipelines in Farsi, Hebrew, Korean, and Turkish.
T5 Model Improvements
- Add 6 new features to T5Transformer for longer and better text generation
- doSample: Whether or not to use sampling; use greedy decoding otherwise
- temperature: The value used to module the next token probabilities
- topK: The number of highest probability vocabulary tokens to keep for top-k-filtering
- topP: If set to float < 1, only the most probable tokens with probabilities that add up to
top_por higher are kept for generation - repetitionPenalty: The parameter for repetition penalty. 1.0 means no penalty. See CTRL: A Conditional Transformer Language Model for Controllable Generation paper for more details
- noRepeatNgramSize: If set to int > 0, all ngrams of that size can only occur once
New Open Source Model in NLU 3.0.2
New multilingual models and pipelines for Farsi, Hebrew, Korean, and Turkish
| Model | NLU Reference | Spark NLP Reference | Lang |
|---|---|---|---|
| ClassifierDLModel | tr.classify.news |
classifierdl_bert_news | tr |
| UniversalSentenceEncoder | xx.use.multi |
tfhub_use_multi | xx |
| UniversalSentenceEncoder | xx.use.multi_lg |
tfhub_use_multi_lg | xx |
| Pipeline | NLU Reference | Spark NLP Reference | Lang |
|---|---|---|---|
| PretrainedPipeline | fa.ner.dl |
recognize_entities_dl | fa |
| PretrainedPipeline | he.explain_document |
explain_document_lg | he |
| PretrainedPipeline | ko.explain_document |
explain_document_lg | ko |
New Healthcare Models in NLU 3.0.2
Five new resolver models:
en.resolve.umls: This model returns CUI (concept unique identifier) codes for Clinical Findings, Medical Devices, Anatomical Structures and Injuries & Poisoning terms.en.resolve.umls.findings: This model returns CUI (concept unique identifier) codes for 200K concepts from clinical findings.en.resolve.loinc: Map clinical NER entities to LOINC codes using sbiobert.en.resolve.loinc.bluebert: Map clinical NER entities to LOINC codes using sbluebert.en.resolve.HPO: This model returns Human Phenotype Ontology (HPO) codes for phenotypic abnormalities encountered in human diseases. It also returns associated codes from the following vocabularies for each HPO code:
| Model | NLU Reference | Spark NLP Reference |
|---|---|---|
| Resolver | en.resolve.umls |
sbiobertresolve_umls_major_concepts |
| Resolver | en.resolve.umls.findings |
sbiobertresolve_umls_findings |
| Resolver | en.resolve.loinc |
sbiobertresolve_loinc |
| Resolver | en.resolve.loinc.biobert |
sbiobertresolve_loinc |
| Resolver | en.resolve.loinc.bluebert |
sbluebertresolve_loinc |
| Resolver | en.resolve.HPO |
sbiobertresolve_HPO |
nlu.load('med_ner.jsl.wip.clinical en.resolve.HPO').viz("""These disorders include cancer, bipolar disorder, schizophrenia, autism, Cri-du-chat syndrome,
myopia, cortical cataract-linked Alzheimer's disease, and infectious diseases""")
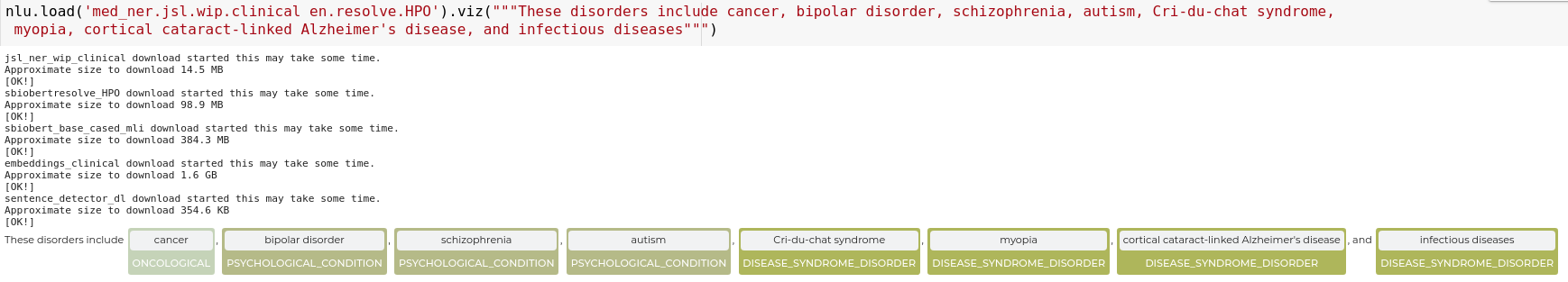
nlu.load('med_ner.jsl.wip.clinical en.resolve.loinc.bluebert').viz("""A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and
subsequent type two diabetes mellitus (TSS2DM), one prior episode of HTG-induced pancreatitis three years prior to presentation, associated with an acute
hepatitis, and obesity with a body mass index (BMI) of 33.5 kg/m2, presented with a one-week history of polyuria, polydipsia, poor appetite, and vomiting.""")
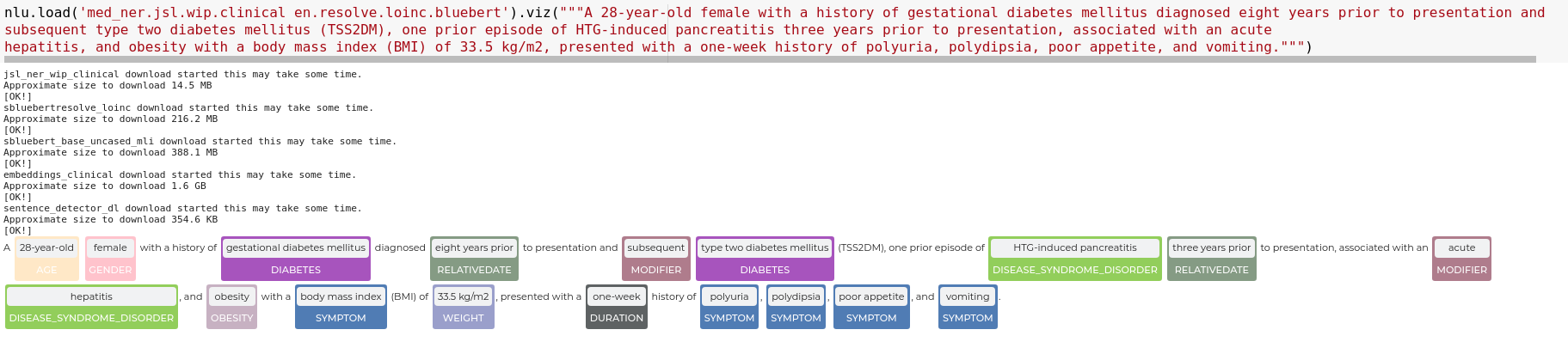
nlu.load('med_ner.jsl.wip.clinical en.resolve.umls.findings').viz("""A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and
subsequent type two diabetes mellitus (TSS2DM), one prior episode of HTG-induced pancreatitis three years prior to presentation, associated with an acute
hepatitis, and obesity with a body mass index (BMI) of 33.5 kg/m2, presented with a one-week history of polyuria, polydipsia, poor appetite, and vomiting."""
)

nlu.load('med_ner.jsl.wip.clinical en.resolve.umls').viz("""A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and
subsequent type two diabetes mellitus (TSS2DM), one prior episode of HTG-induced pancreatitis three years prior to presentation, associated with an acute
hepatitis, and obesity with a body mass index (BMI) of 33.5 kg/m2, presented with a one-week history of polyuria, polydipsia, poor appetite, and vomiting.""")
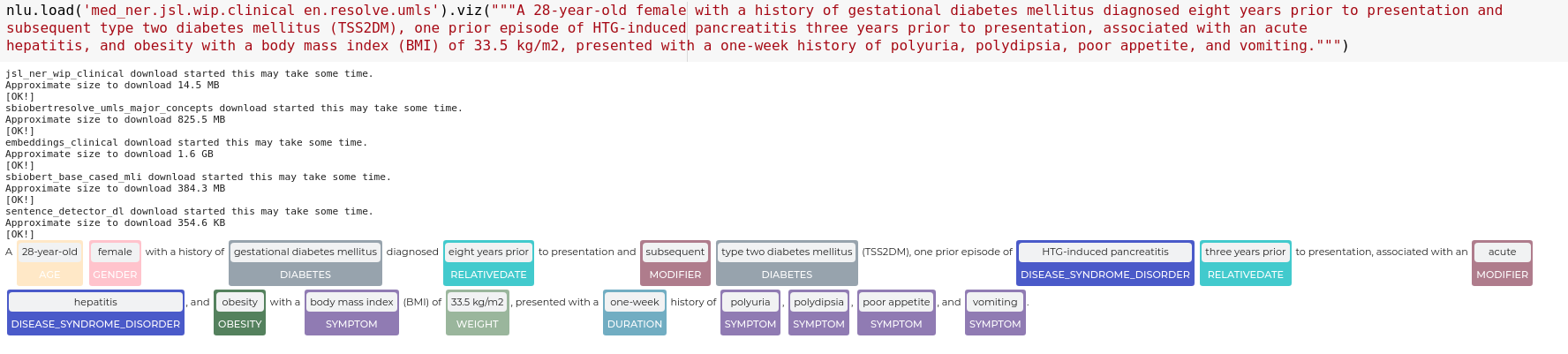
nlu.load('med_ner.jsl.wip.clinical en.resolve.loinc').predict("""A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and
subsequent type two diabetes mellitus (TSS2DM), one prior episode of HTG-induced pancreatitis three years prior to presentation, associated with an acute
hepatitis, and obesity with a body mass index (BMI) of 33.5 kg/m2, presented with a one-week history of polyuria, polydipsia, poor appetite, and vomiting.""")
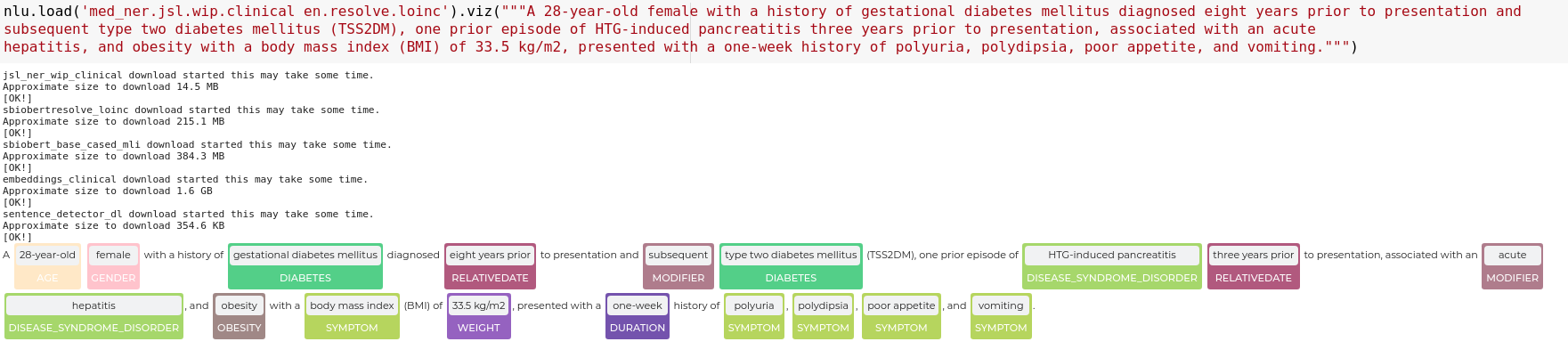
nlu.load('med_ner.jsl.wip.clinical en.resolve.loinc.biobert').predict("""A 28-year-old female with a history of gestational diabetes mellitus diagnosed eight years prior to presentation and
subsequent type two diabetes mellitus (TSS2DM), one prior episode of HTG-induced pancreatitis three years prior to presentation, associated with an acute
hepatitis, and obesity with a body mass index (BMI) of 33.5 kg/m2, presented with a one-week history of polyuria, polydipsia, poor appetite, and vomiting.""")

- 140+ tutorials
- New Streamlit visualizations docs
- The complete list of all 1100+ models & pipelines in 192+ languages is available on Models Hub.
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : !wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark==3.0.1
NLU Version 3.0.1
We are very excited to announce NLU 3.0.1 has been released!
This is one of the most visually appealing releases, with the integration of the Spark-NLP-Display library and visualizations for dependency trees, entity resolution, entity assertion, relationship between entities and named
entity recognition. In addition to this, the schema of how columns are named by NLU has been reworked and all 140+ tutorial notebooks have been updated to reflect the latest changes in NLU 3.0.0+
Finally, new multilingual models for Afrikaans, Welsh, Maltese, Tamil, andVietnamese are now available.
New Features and Enhancements
- 1 line to visualization for
NER,Dependency,Resolution,AssertionandRelationvia Spark-NLP-Display integration - Improved column naming schema
- Over 140 + NLU tutorial Notebooks updated and improved to reflect latest changes in NLU 3.0.0 +
- New multilingual models for
Afrikaans,Welsh,Maltese,Tamil, andVietnamese
Improved Column Name generation
- NLU categorized each internal component now with boolean labels for
name_deductableandalways_name_deductable. - Before generating column names, NLU checks wether each component is of unique in the pipeline or not. If a component is not unique in the
pipe and there are multiple components of same type, i.e. multiple
NERmodels, NLU will deduct a base name for the final output columns from the NLU reference each NER model is pointing to. - If on the other hand, there is only one
NERmodel in the pipeline, only the defaultnercolumn prefixed will be generated. - For some components, like
embeddingsandclassifiersare now defined asalways_name_deductable, for those NLU will always try to infer a meaningful base name for the output columns. - Newly trained component output columns will now be prefixed with
trained_<type>, for typespos,ner,cLassifier,sentimentandmulti_classifier
Enhanced offline mode
- You can still load a model from a path as usual with
nlu.load(path=model_path)and output columns will be suffixed withfrom_disk - You can now optionally also specify
requestparameter during load a model from HDD, it will be used to deduct more meaningful column name suffixes, instead offrom_disk, i.e. by callingnlu.load(request ='en.embed_sentence.biobert.pubmed_pmc_base_cased', path=model_path)
NLU visualization
The latest NLU release integrated the beautiful Spark-NLP-Display package visualizations. You do not need to worry about installing it, when you try to visualize something, NLU will check if Spark-NLP-Display is installed, if it is missing it will be dynamically installed into your python executable environment, so you don’t need to worry about anything!
See the visualization tutorial notebook and visualization docs for more info.
NER visualization
Applicable to any of the 100+ NER models! See here for an overview
nlu.load('ner').viz("Donald Trump from America and Angela Merkel from Germany don't share many oppinions.")

Dependency tree visualization
Visualizes the structure of the labeled dependency tree and part of speech tags
nlu.load('dep.typed').viz("Billy went to the mall")
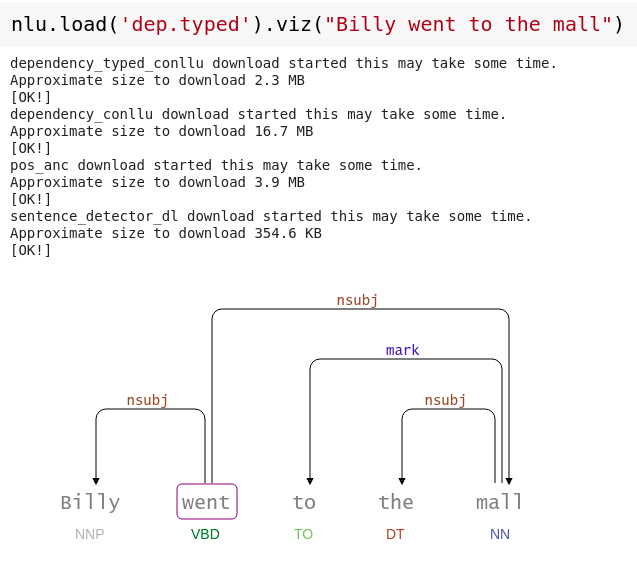
#Bigger Example
nlu.load('dep.typed').viz("Donald Trump from America and Angela Merkel from Germany don't share many oppinions but they both love John Snow Labs software")
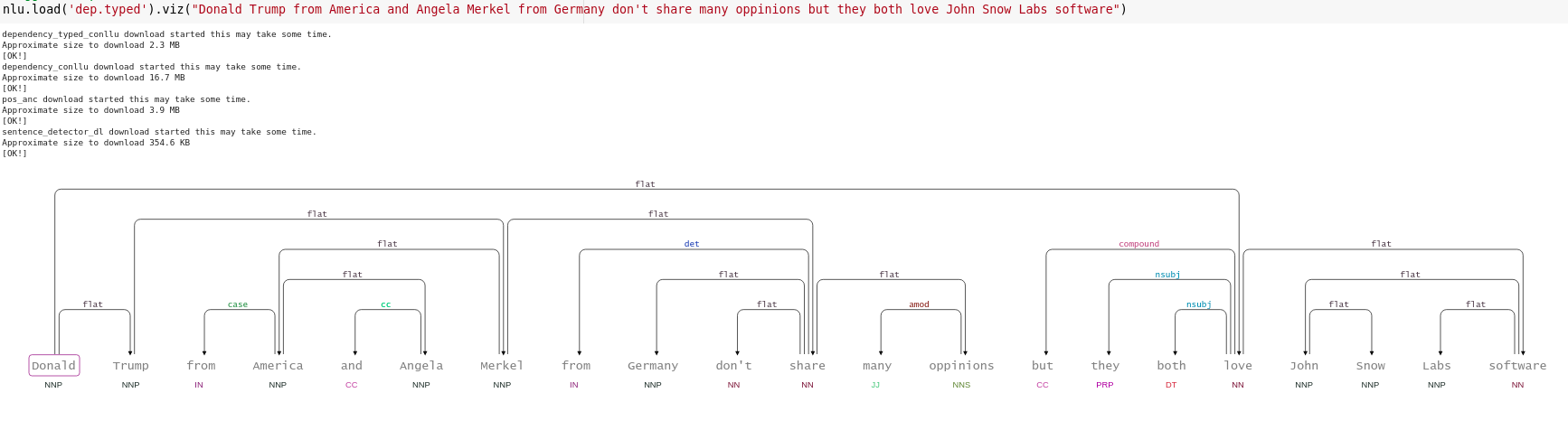
Assertion status visualization
Visualizes asserted statuses and entities.
Applicable to any of the 10 + Assertion models! See here for an overview
nlu.load('med_ner.clinical assert').viz("The MRI scan showed no signs of cancer in the left lung")
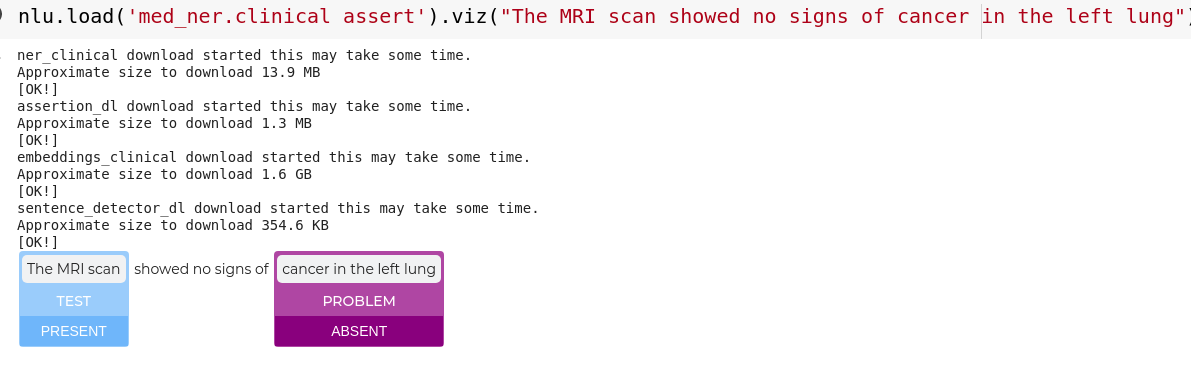
#bigger example
data ='This is the case of a very pleasant 46-year-old Caucasian female, seen in clinic on 12/11/07 during which time MRI of the left shoulder showed no evidence of rotator cuff tear. She did have a previous MRI of the cervical spine that did show an osteophyte on the left C6-C7 level. Based on this, negative MRI of the shoulder, the patient was recommended to have anterior cervical discectomy with anterior interbody fusion at C6-C7 level. Operation, expected outcome, risks, and benefits were discussed with her. Risks include, but not exclusive of bleeding and infection, bleeding could be soft tissue bleeding, which may compromise airway and may result in return to the operating room emergently for evacuation of said hematoma. There is also the possibility of bleeding into the epidural space, which can compress the spinal cord and result in weakness and numbness of all four extremities as well as impairment of bowel and bladder function. However, the patient may develop deeper-seated infection, which may require return to the operating room. Should the infection be in the area of the spinal instrumentation, this will cause a dilemma since there might be a need to remove the spinal instrumentation and/or allograft. There is also the possibility of potential injury to the esophageus, the trachea, and the carotid artery. There is also the risks of stroke on the right cerebral circulation should an undiagnosed plaque be propelled from the right carotid. She understood all of these risks and agreed to have the procedure performed.'
nlu.load('med_ner.clinical assert').viz(data)
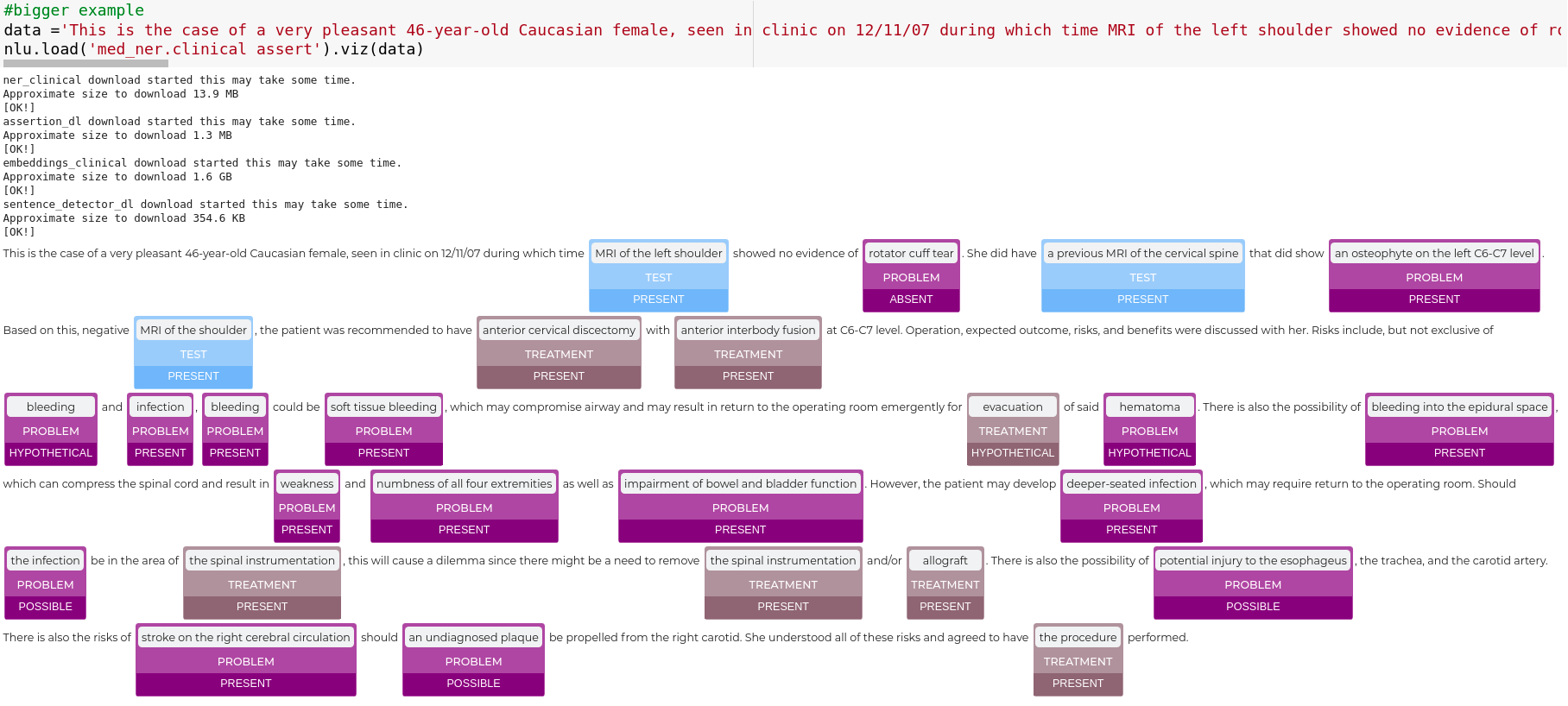
Relationship between entities visualization
Visualizes the extracted entities between relationship.
Applicable to any of the 20 + Relation Extractor models See here for an overview
nlu.load('med_ner.jsl.wip.clinical relation.temporal_events').viz('The patient developed cancer after a mercury poisoning in 1999 ')

# bigger example
data = 'This is the case of a very pleasant 46-year-old Caucasian female, seen in clinic on 12/11/07 during which time MRI of the left shoulder showed no evidence of rotator cuff tear. She did have a previous MRI of the cervical spine that did show an osteophyte on the left C6-C7 level. Based on this, negative MRI of the shoulder, the patient was recommended to have anterior cervical discectomy with anterior interbody fusion at C6-C7 level. Operation, expected outcome, risks, and benefits were discussed with her. Risks include, but not exclusive of bleeding and infection, bleeding could be soft tissue bleeding, which may compromise airway and may result in return to the operating room emergently for evacuation of said hematoma. There is also the possibility of bleeding into the epidural space, which can compress the spinal cord and result in weakness and numbness of all four extremities as well as impairment of bowel and bladder function. However, the patient may develop deeper-seated infection, which may require return to the operating room. Should the infection be in the area of the spinal instrumentation, this will cause a dilemma since there might be a need to remove the spinal instrumentation and/or allograft. There is also the possibility of potential injury to the esophageus, the trachea, and the carotid artery. There is also the risks of stroke on the right cerebral circulation should an undiagnosed plaque be propelled from the right carotid. She understood all of these risks and agreed to have the procedure performed'
pipe = nlu.load('med_ner.jsl.wip.clinical relation.clinical').viz(data)
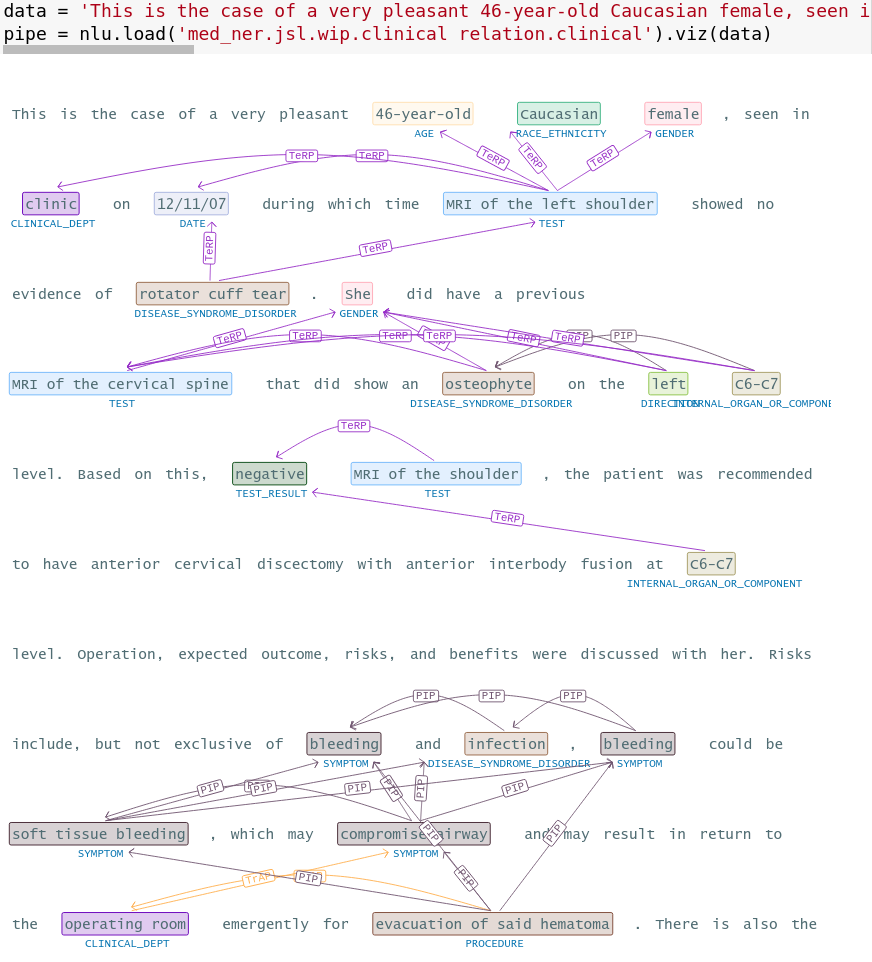
Entity Resolution visualization for chunks
Visualizes resolutions of entities Applicable to any of the 100+ Resolver models See here for an overview
nlu.load('med_ner.jsl.wip.clinical resolve_chunk.rxnorm.in').viz("He took Prevacid 30 mg daily")

# bigger example
data = "This is an 82 - year-old male with a history of prior tobacco use , hypertension , chronic renal insufficiency , COPD , gastritis , and TIA who initially presented to Braintree with a non-ST elevation MI and Guaiac positive stools , transferred to St . Margaret\'s Center for Women & Infants for cardiac catheterization with PTCA to mid LAD lesion complicated by hypotension and bradycardia requiring Atropine , IV fluids and transient dopamine possibly secondary to vagal reaction , subsequently transferred to CCU for close monitoring , hemodynamically stable at the time of admission to the CCU ."
nlu.load('med_ner.jsl.wip.clinical resolve_chunk.rxnorm.in').viz(data)
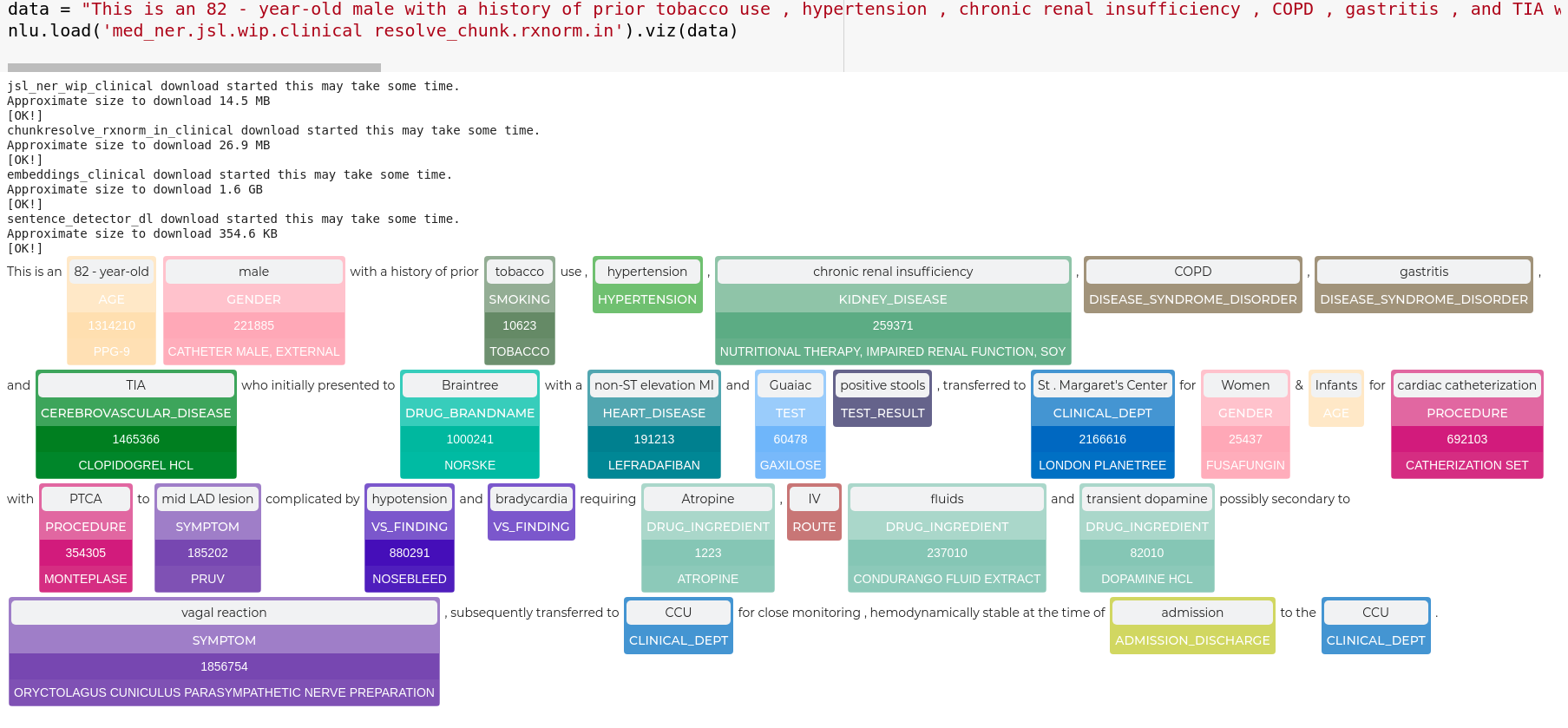
Entity Resolution visualization for sentences
Visualizes resolutions of entities in sentences Applicable to any of the 100+ Resolver models See here for an overview
nlu.load('med_ner.jsl.wip.clinical resolve.icd10cm').viz('She was diagnosed with a respiratory congestion')

# bigger example
data = 'The patient is a 5-month-old infant who presented initially on Monday with a cold, cough, and runny nose for 2 days. Mom states she had no fever. Her appetite was good but she was spitting up a lot. She had no difficulty breathing and her cough was described as dry and hacky. At that time, physical exam showed a right TM, which was red. Left TM was okay. She was fairly congested but looked happy and playful. She was started on Amoxil and Aldex and we told to recheck in 2 weeks to recheck her ear. Mom returned to clinic again today because she got much worse overnight. She was having difficulty breathing. She was much more congested and her appetite had decreased significantly today. She also spiked a temperature yesterday of 102.6 and always having trouble sleeping secondary to congestion'
nlu.load('med_ner.jsl.wip.clinical resolve.icd10cm').viz(data)
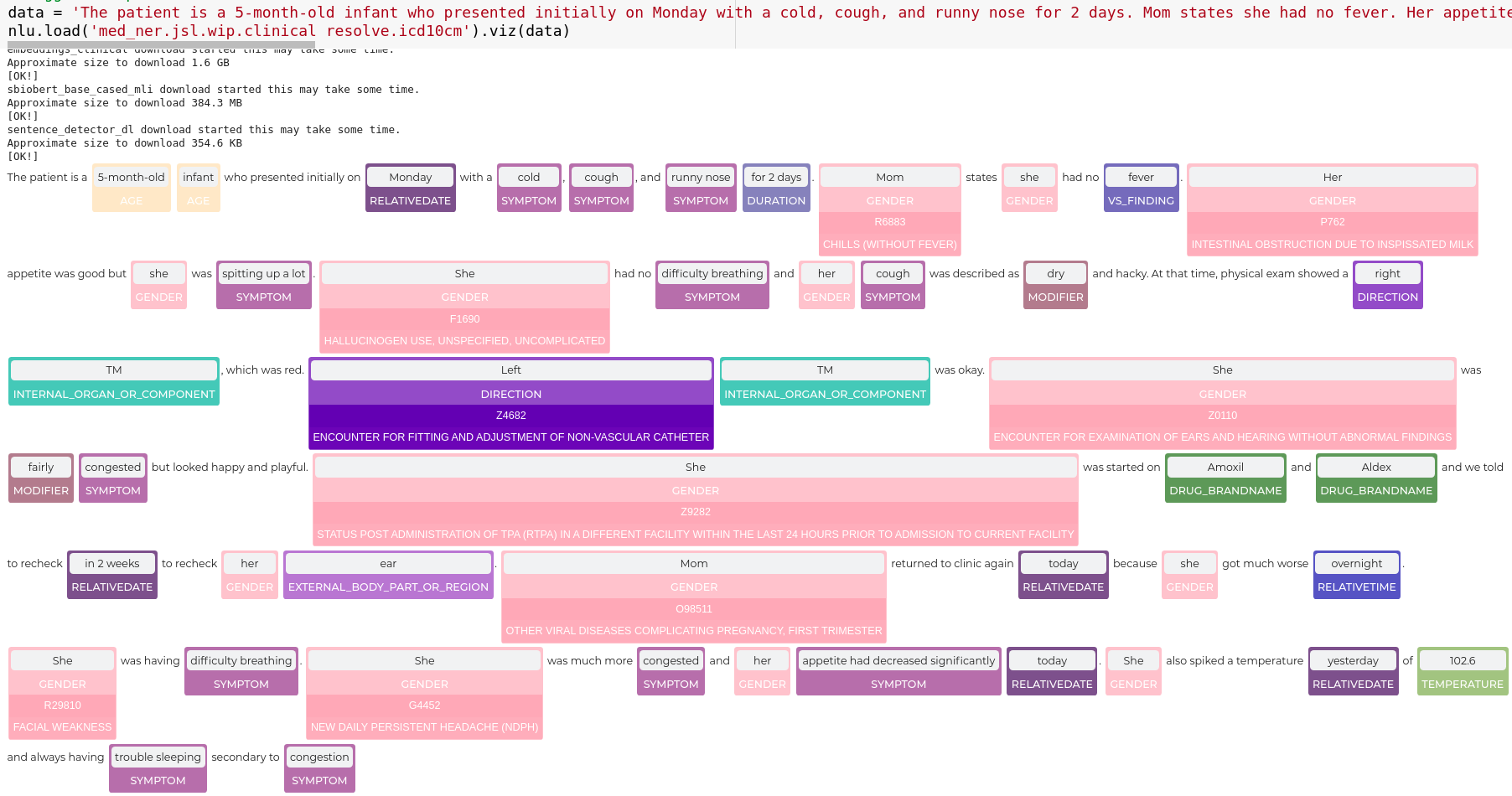
Configure visualizations
Define custom colors for labels
Some entity and relation labels will be highlighted with a pre-defined color, which you can find here.
For labels that have no color defined, a random color will be generated.
You can define colors for labels manually, by specifying via the viz_colors parameter
and defining hex color codes in a dictionary that maps labels to colors .
data = 'Dr. John Snow suggested that Fritz takes 5mg penicilin for his cough'
# Define custom colors for labels
viz_colors={'STRENGTH':'#800080', 'DRUG_BRANDNAME':'#77b5fe', 'GENDER':'#77ffe'}
nlu.load('med_ner.jsl.wip.clinical').viz(data,viz_colors =viz_colors)
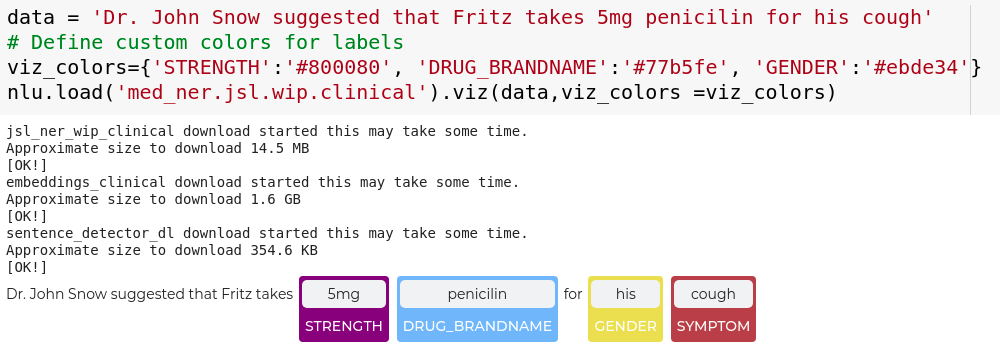
Filter entities that get highlighted
By default every entity class will be visualized.
The labels_to_viz can be used to define a set of labels to highlight.
Applicable for ner, resolution and assert.
data = 'Dr. John Snow suggested that Fritz takes 5mg penicilin for his cough'
# Filter wich NER label to viz
labels_to_viz=['SYMPTOM']
nlu.load('med_ner.jsl.wip.clinical').viz(data,labels_to_viz=labels_to_viz)

New models
New multilingual models for Afrikaans, Welsh, Maltese, Tamil, andVietnamese
| nlu.load() Refrence | Spark NLP Refrence |
|---|---|
| vi.lemma | lemma |
| mt.lemma | lemma |
| ta.lemma | lemma |
| af.lemma | lemma |
| af.pos | pos_afribooms |
| cy.lemma | lemma |
Reworked and updated NLU tutorial notebooks
All of the 140+ NLU tutorial Notebooks have been updated and reworked to reflect the latest changes in NLU 3.0.0+
Bugfixes
- Fixed a bug that caused resolution algorithms output level to be inferred incorrectly
- Fixed a bug that caused stranger cols got dropped
- Fixed a bug that caused endings to miss when .predict(position=True) was specified
- Fixed a bug that caused pd.Series to be converted incorrectly internally
- Fixed a bug that caused output level transformations to crash
- Fixed a bug that caused verbose mode not to turn of properly after turning it on.
-
fixed a bug that caused some models to crash when loaded for HDD
- 140+ updates tutorials
- Updated visualization docs
- Models Hub with new models
- Spark NLP publications
- NLU in Action
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
Install NLU in 1 line!aaa
* Install NLU on Google Colab : ! wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU on Kaggle : ! wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark==3.0.3
200+ State of the Art Medical Models for NER, Entity Resolution, Relation Extraction, Assertion, Spark 3 and Python 3.8 support in NLU 3.0 Release and much more
We are incredible excited to announce the release of NLU 3.0.0 which makes most of John Snow Labs medical healthcare model available in just 1 line of code in NLU.
These models are the most accurate in their domains and highly scalable in Spark clusters.
In addition, Spark 3.0.X and Spark 3.1.X is now supported, together with Python3.8
This is enabled by the the amazing Spark NLP3.0.1 and Spark NLP for Healthcare 3.0.1 releases.
New Features
- Over 200 new models for the
healthcaredomain - 6 new classes of models, Assertion, Sentence/Chunk Resolvers, Relation Extractors, Medical NER models, De-Identificator Models
- Spark 3.0.X and 3.1.X support
- Python 3.8 Support
- New Output level
relation - 1 Line to install NLU just run
!wget https://raw.githubusercontent.com/JohnSnowLabs/nlu/master/scripts/colab_setup.sh -O - | bash - Various new EMR and Databricks versions supported
- GPU Mode, more then 600% speedup by enabling GPU mode.
- Authorized mode for licensed features
New Documentation
New Notebooks
- Medical Named Entity Extraction (NER) notebook
- Relation extraction notebook
- Entity Resolution overview notebook
- Assertion overview notebook
- De-Identification overview notebook
- Graph NLU tutorial for the GRAPH+AI Summit hosted by Tigergraph
AssertionDLModels
| Language | nlu.load() reference | Spark NLP Model reference |
|---|---|---|
| English | assert | assertion_dl |
| English | assert.biobert | assertion_dl_biobert |
| English | assert.healthcare | assertion_dl_healthcare |
| English | assert.large | assertion_dl_large |
New Word Embeddings
| Language | nlu.load() reference | Spark NLP Model reference |
|---|---|---|
| English | embed.glove.clinical | embeddings_clinical |
| English | embed.glove.biovec | embeddings_biovec |
| English | embed.glove.healthcare | embeddings_healthcare |
| English | embed.glove.healthcare_100d | embeddings_healthcare_100d |
| English | en.embed.glove.icdoem | embeddings_icdoem |
| English | en.embed.glove.icdoem_2ng | embeddings_icdoem_2ng |
Sentence Entity resolvers
RelationExtractionModel
| Language | nlu.load() reference | Spark NLP Model reference |
|---|---|---|
| English | relation.posology | posology_re |
| English | relation | redl_bodypart_direction_biobert |
| English | relation.bodypart.direction | redl_bodypart_direction_biobert |
| English | relation.bodypart.problem | redl_bodypart_problem_biobert |
| English | relation.bodypart.procedure | redl_bodypart_procedure_test_biobert |
| English | relation.chemprot | redl_chemprot_biobert |
| English | relation.clinical | redl_clinical_biobert |
| English | relation.date | redl_date_clinical_biobert |
| English | relation.drug_drug_interaction | redl_drug_drug_interaction_biobert |
| English | relation.humen_phenotype_gene | redl_human_phenotype_gene_biobert |
| English | relation.temporal_events | redl_temporal_events_biobert |
NERDLModels
De-Identification Models
| Language | nlu.load() reference | Spark NLP Model reference |
|---|---|---|
| English | med_ner.deid.augmented | ner_deid_augmented |
| English | med_ner.deid.biobert | ner_deid_biobert |
| English | med_ner.deid.enriched | ner_deid_enriched |
| English | med_ner.deid.enriched_biobert | ner_deid_enriched_biobert |
| English | med_ner.deid.large | ner_deid_large |
| English | med_ner.deid.sd | ner_deid_sd |
| English | med_ner.deid.sd_large | ner_deid_sd_large |
| English | med_ner.deid | nerdl_deid |
| English | med_ner.deid.synthetic | ner_deid_synthetic |
| English | med_ner.deid.dl | ner_deidentify_dl |
| English | en.de_identify | deidentify_rb |
| English | de_identify.rules | deid_rules |
| English | de_identify.clinical | deidentify_enriched_clinical |
| English | de_identify.large | deidentify_large |
| English | de_identify.rb | deidentify_rb |
| English | de_identify.rb_no_regex | deidentify_rb_no_regex |
Chunk resolvers
New Classifiers
| Language | nlu.load() reference | Spark NLP Model reference |
|---|---|---|
| English | classify.icd10.clinical | classifier_icd10cm_hcc_clinical |
| English | classify.icd10.healthcare | classifier_icd10cm_hcc_healthcare |
| English | classify.ade.biobert | classifierdl_ade_biobert |
| English | classify.ade.clinical | classifierdl_ade_clinicalbert |
| English | classify.ade.conversational | classifierdl_ade_conversational_biobert |
| English | classify.gender.biobert | classifierdl_gender_biobert |
| English | classify.gender.sbert | classifierdl_gender_sbert |
| English | classify.pico | classifierdl_pico_biobert |
German Medical models
| nlu.load() reference | Spark NLP Model reference |
|---|---|
| [embed] | w2v_cc_300d |
| [embed.w2v] | w2v_cc_300d |
| [resolve_chunk] | chunkresolve_ICD10GM |
| [resolve_chunk.icd10gm] | chunkresolve_ICD10GM |
| resolve_chunk.icd10gm.2021 | chunkresolve_ICD10GM_2021 |
| med_ner.legal | ner_legal |
| med_ner | ner_healthcare |
| med_ner.healthcare | ner_healthcare |
| med_ner.healthcare_slim | ner_healthcare_slim |
| med_ner.traffic | ner_traffic |
Spanish Medical models
GPU Mode
You can now enable NLU GPU mode by setting gpu=true while loading a model. I.e. nlu.load('train.sentiment' gpu=True) . If must resart you kernel, if you already loaded a nlu pipeline withouth GPU mode.
Output Level Relation
This new output level is used for relation extractors and will give you 1 row per relation extracted.
Bug fixes
- Fixed a bug that caused loading NLU models in offline mode not to work in some occasions
Install NLU in 1 line!
* Install NLU on Google Colab : !wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
* Install NLU via Pip : ! pip install nlu pyspark==3.0.3
Additional NLU ressources
NLU Version 1.1.3
Intent and Action Classification, analyze Chinese News and the Crypto market, train a classifier that understands 100+ languages, translate between 200 + languages, answer questions, summarize text, and much more in NLU 1.1.3
We are very excited to announce that the latest NLU release comes with a new pretrained Intent Classifier and NER Action Extractor for text related to music, restaurants, and movies trained on the SNIPS dataset. Make sure to check out the models hub and the easy 1-liners for more info!
In addition to that, new NER and Embedding models for Bengali are now available
Finally, there is a new NLU Webinar with 9 accompanying tutorial notebooks which teach you a lot of things and is segmented into the following parts :
- Part1: Easy 1 Liners
- Spell checking/Sentiment/POS/NER/ BERTtology embeddings
- Part2: Data analysis and NLP tasks on Crypto News Headline dataset
- Preprocessing and extracting Emotions, Keywords, Named Entities and visualize them
- Part3: NLU Multi-Lingual 1 Liners with Microsoft’s Marian Models
- Translate between 200+ languages (and classify lang afterward)
- Part 4: Data analysis and NLP tasks on Chinese News Article Dataset
- Word Segmentation, Lemmatization, Extract Keywords, Named Entities and translate to english
- Part 5: Train a sentiment Classifier that understands 100+ Languages
- Train on a french sentiment dataset and predict the sentiment of 100+ languages with language-agnostic BERT Sentence Embedding
- Part 6: Question answering, Summarization, Squad and more with Google’s T5
- T5 Question answering and 18 + other NLP tasks (SQUAD / GLUE / SUPER GLUE)
New Models
NLU 1.1.3 New Non-English Models
| Language | nlu.load() reference | Spark NLP Model reference | Type |
|---|---|---|---|
| Bengali | bn.ner.cc_300d | bengaliner_cc_300d | NerDLModel |
| Bengali | bn.embed | bengali_cc_300d | NerDLModel |
| Bengali | bn.embed.cc_300d | bengali_cc_300d | Word Embeddings Model (Alias) |
| Bengali | bn.embed.glove | bengali_cc_300d | Word Embeddings Model (Alias) |
NLU 1.1.3 New English Models
| Language | nlu.load() reference | Spark NLP Model reference | Type |
|---|---|---|---|
| English | en.classify.snips | nerdl_snips_100d | NerDLModel |
| English | en.ner.snips | classifierdl_use_snips | ClassifierDLModel |
New NLU Webinar
State-of-the-art Natural Language Processing for 200+ Languages with 1 Line of code
Talk Abstract
Learn to harness the power of 1,000+ production-grade & scalable NLP models for 200+ languages - all available with just 1 line of Python code by leveraging the open-source NLU library, which is powered by the widely popular Spark NLP.
John Snow Labs has delivered over 80 releases of Spark NLP to date, making it the most widely used NLP library in the enterprise and providing the AI community with state-of-the-art accuracy and scale for a variety of common NLP tasks. The most recent releases include pre-trained models for over 200 languages - including languages that do not use spaces for word segmentation algorithms like Chinese, Japanese, and Korean, and languages written from right to left like Arabic, Farsi, Urdu, and Hebrew. All software and models are free and open source under an Apache 2.0 license.
This webinar will show you how to leverage the multi-lingual capabilities of Spark NLP & NLU - including automated language detection for up to 375 languages, and the ability to perform translation, named entity recognition, stopword removal, lemmatization, and more in a variety of language families. We will create Python code in real-time and solve these problems in just 30 minutes. The notebooks will then be made freely available online.
You can watch the video here,
NLU 1.1.3 New Notebooks and tutorials
New Webinar Notebooks
- NLU basics, easy 1-liners (Spellchecking, sentiment, NER, POS, BERT
- Analyze Crypto News dataset with Keyword extraction, NER, Emotional distribution, and stemming
- Translate Crypto News dataset between 300 Languages with the Marian Model (German, French, Hebrew examples)
- Translate Crypto News dataset between 300 Languages with the Marian Model (Hindi, Russian, Chinese examples)
- Analyze Chinese News Headlines with Chinese Word Segmentation, Lemmatization, NER, and Keyword extraction
- Train a Sentiment Classifier that will understand 100+ languages on just a French Dataset with the powerful Language Agnostic Bert Embeddings
- Summarize text and Answer Questions with T5
- Solve any task in 1 line from SQUAD, GLUE and SUPER GLUE with T5
- Overview of models for various languages
New easy NLU 1-liners in NLU 1.1.3
Detect actions in general commands related to music, restaurant, movies.
nlu.load("en.classify.snips").predict("book a spot for nona gray myrtle and alison at a top-rated brasserie that is distant from wilson av on nov the 4th 2030 that serves ouzeri",output_level = "document")
outputs :
| ner_confidence | entities | document | Entities_Classes |
|---|---|---|---|
| [1.0, 1.0, 0.9997000098228455, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 0.9990000128746033, 1.0, 1.0, 1.0, 0.9965000152587891, 0.9998999834060669, 0.9567000269889832, 1.0, 1.0, 1.0, 0.9980000257492065, 0.9991999864578247, 0.9988999962806702, 1.0, 1.0, 0.9998999834060669] | [‘nona gray myrtle and alison’, ‘top-rated’, ‘brasserie’, ‘distant’, ‘wilson av’, ‘nov the 4th 2030’, ‘ouzeri’] | book a spot for nona gray myrtle and alison at a top-rated brasserie that is distant from wilson av on nov the 4th 2030 that serves ouzeri | [‘party_size_description’, ‘sort’, ‘restaurant_type’, ‘spatial_relation’, ‘poi’, ‘timeRange’, ‘cuisine’] |
Named Entity Recognition (NER) Model in Bengali (bengaliner_cc_300d)
# Bengali for: 'Iajuddin Ahmed passed Matriculation from Munshiganj High School in 1947 and Intermediate from Munshiganj Horganga College in 1950.'
nlu.load("bn.ner.cc_300d").predict("১৯৪৮ সালে ইয়াজউদ্দিন আহম্মেদ মুন্সিগঞ্জ উচ্চ বিদ্যালয় থেকে মেট্রিক পাশ করেন এবং ১৯৫০ সালে মুন্সিগঞ্জ হরগঙ্গা কলেজ থেকে ইন্টারমেডিয়েট পাশ করেন",output_level = "document")
outputs :
| ner_confidence | entities | Entities_Classes | document |
|---|---|---|---|
| [0.9987999796867371, 0.9854000210762024, 0.8604000210762024, 0.6686999797821045, 0.5289999842643738, 0.7009999752044678, 0.7684999704360962, 0.9979000091552734, 0.9976000189781189, 0.9930999875068665, 0.9994000196456909, 0.9879000186920166, 0.7407000064849854, 0.9215999841690063, 0.7657999992370605, 0.39419999718666077, 0.9124000072479248, 0.9932000041007996, 0.9919999837875366, 0.995199978351593, 0.9991999864578247] | [‘সালে’, ‘ইয়াজউদ্দিন আহম্মেদ’, ‘মুন্সিগঞ্জ উচ্চ বিদ্যালয়’, ‘সালে’, ‘মুন্সিগঞ্জ হরগঙ্গা কলেজ’] | [‘TIME’, ‘PER’, ‘ORG’, ‘TIME’, ‘ORG’] | ১৯৪৮ সালে ইয়াজউদ্দিন আহম্মেদ মুন্সিগঞ্জ উচ্চ বিদ্যালয় থেকে মেট্রিক পাশ করেন এবং ১৯৫০ সালে মুন্সিগঞ্জ হরগঙ্গা কলেজ থেকে ইন্টারমেডিয়েট পাশ করেন |
Identify intent in general text - SNIPS dataset
nlu.load("en.ner.snips").predict("I want to bring six of us to a bistro in town that serves hot chicken sandwich that is within the same area",output_level = "document")
outputs :
| document | snips | snips_confidence |
|---|---|---|
| I want to bring six of us to a bistro in town that serves hot chicken sandwich that is within the same area | BookRestaurant | 1 |
Word Embeddings for Bengali (bengali_cc_300d)
# Bengali for : 'Iajuddin Ahmed passed Matriculation from Munshiganj High School in 1947 and Intermediate from Munshiganj Horganga College in 1950.'
nlu.load("bn.embed").predict("১৯৪৮ সালে ইয়াজউদ্দিন আহম্মেদ মুন্সিগঞ্জ উচ্চ বিদ্যালয় থেকে মেট্রিক পাশ করেন এবং ১৯৫০ সালে মুন্সিগঞ্জ হরগঙ্গা কলেজ থেকে ইন্টারমেডিয়েট পাশ করেন",output_level = "document")
outputs :
| document | bn_embed_embeddings |
|---|---|
| ১৯৪৮ সালে ইয়াজউদ্দিন আহম্মেদ মুন্সিগঞ্জ উচ্চ বিদ্যালয় থেকে মেট্রিক পাশ করেন এবং ১৯৫০ সালে মুন্সিগঞ্জ হরগঙ্গা কলেজ থেকে ইন্টারমেডিয়েট পাশ করেন | [-0.0828 0.0683 0.0215 … 0.0679 -0.0484…] |
NLU 1.1.3 Enhancements
- Added automatic conversion to Sentence Embeddings of Word Embeddings when there is no Sentence Embedding Avaiable and a model needs the converted version to run.
NLU 1.1.3 Bug Fixes
- Fixed a bug that caused
ur.sentimentNLU pipeline to build incorrectly - Fixed a bug that caused
sentiment.imdb.gloveNLU pipeline to build incorrectly - Fixed a bug that caused
en.sentiment.glove.imdbNLU pipeline to build incorrectly - Fixed a bug that caused Spark 2.3.X environments to crash.
NLU Installation
# PyPi
!pip install nlu pyspark==2.4.7
#Conda
# Install NLU from Anaconda/Conda
conda install -os_components johnsnowlabs nlu
Additional NLU ressources
NLU Version 1.1.2
Hindi WordEmbeddings , Bengali Named Entity Recognition (NER), 30+ new models, analyze Crypto news with John Snow Labs NLU 1.1.2
We are very happy to announce NLU 1.1.2 has been released with the integration of 30+ models and pipelines Bengali Named Entity Recognition, Hindi Word Embeddings,
and state-of-the-art transformer based OntoNotes models and pipelines from the incredible Spark NLP 2.7.3 Release in addition to a few bugfixes.
In addition to that, there is a new NLU Webinar video showcasing in detail
how to use NLU to analyze a crypto news dataset to extract keywords unsupervised and predict sentimential/emotional distributions of the dataset and much more!
Python’s NLU library: 1,000+ models, 200+ Languages, State of the Art Accuracy, 1 Line of code - NLU NYC/DC NLP Meetup Webinar
Using just 1 line of Python code by leveraging the NLU library, which is powered by the award-winning Spark NLP.
This webinar covers, using live coding in real-time, how to deliver summarization, translation, unsupervised keyword extraction, emotion analysis, question answering, spell checking, named entity recognition, document classification, and other common NLP tasks. T his is all done with a single line of code, that works directly on Python strings or pandas data frames. Since NLU is based on Spark NLP, no code changes are required to scale processing to multi-core or cluster environment - integrating natively with Ray, Dask, or Spark data frames.
The recent releases for Spark NLP and NLU include pre-trained models for over 200 languages and language detection for 375 languages. This includes 20 languages families; non-Latin alphabets; languages that do not use spaces for word segmentation like Chinese, Japanese, and Korean; and languages written from right to left like Arabic, Farsi, Urdu, and Hebrew. We’ll also cover some of the algorithms and models that are included. The code notebooks will be freely available online.
NLU 1.1.2 New Non-English Models
| Language | nlu.load() reference | Spark NLP Model reference | Type |
|---|---|---|---|
| Bengali | bn.ner | ner_jifs_glove_840B_300d | Word Embeddings Model (Alias) |
| Bengali | bn.ner.glove | ner_jifs_glove_840B_300d | Word Embeddings Model (Alias) |
| Hindi | hi.embed | hindi_cc_300d | NerDLModel |
| Bengali | bn.lemma | lemma | Lemmatizer |
| Japanese | ja.lemma | lemma | Lemmatizer |
| Bihari | bh.lemma | lemma | Lemma |
| Amharic | am.lemma | lemma | Lemma |
NLU 1.1.2 New English Models and Pipelines
New Tutorials and Notebooks
- NYC/DC NLP Meetup Webinar video analyze Crypto News, Unsupervised Keywords, Translate between 300 Languages, Question Answering, Summerization, POS, NER in 1 line of code in almost just 20 minutes
- NLU basics POS/NER/Sentiment Classification/BERTology Embeddings
- Explore Crypto Newsarticle dataset, unsupervised Keyword extraction, Stemming, Emotion/Sentiment distribution Analysis
- Translate between more than 300 Languages in 1 line of code with the Marian Models
- New NLU 1.1.2 Models Showcase Notebooks, Bengali NER, Hindi Embeddings, 30 new_models
NLU 1.1.2 Bug Fixes
- Fixed a bug that caused NER confidences not beeing extracted
- Fixed a bug that caused nlu.load(‘spell’) to crash
- Fixed a bug that caused Uralic/Estonian/ET language models not to be loaded properly
New Easy NLU 1-liners in 1.1.2
Named Entity Recognition for Bengali (GloVe 840B 300d)
#Bengali for : It began to be widely used in the United States in the early '90s.
nlu.load("bn.ner").predict("৯০ এর দশকের শুরুর দিকে বৃহৎ আকারে মার্কিন যুক্তরাষ্ট্রে এর প্রয়োগের প্রক্রিয়া শুরু হয়'")
output :
| entities | token | Entities_classes | ner_confidence |
|---|---|---|---|
| [‘মার্কিন যুক্তরাষ্ট্রে’] | ৯০ | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | এর | [‘LOC’] | 0.9999 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | দশকের | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | শুরুর | [‘LOC’] | 0.9969 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | দিকে | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | বৃহৎ | [‘LOC’] | 0.9994 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | আকারে | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | মার্কিন | [‘LOC’] | 0.9602 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | যুক্তরাষ্ট্রে | [‘LOC’] | 0.4134 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | এর | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | প্রয়োগের | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | প্রক্রিয়া | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | শুরু | [‘LOC’] | 0.9999 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | হয় | [‘LOC’] | 1 |
| [‘মার্কিন যুক্তরাষ্ট্রে’] | ’ | [‘LOC’] | 1 |
Bengali Lemmatizer
#Bengali for : One morning in the marble-decorated building of Vaidyanatha, an obese monk was engaged in the enchantment of Duis and the milk service of one and a half Vaidyanatha. Give me two to eat
nlu.load("bn.lemma").predict("একদিন প্রাতে বৈদ্যনাথের মার্বলমণ্ডিত দালানে একটি স্থূলোদর সন্ন্যাসী দুইসের মোহনভোগ এবং দেড়সের দুগ্ধ সেবায় নিযুক্ত আছে বৈদ্যনাথ গায়ে একখানি চাদর দিয়া জোড়করে একান্ত বিনীতভাবে ভূতলে বসিয়া ভক্তিভরে পবিত্র ভোজনব্যাপার নিরীক্ষণ করিতেছিলেন এমন সময় কোনোমতে দ্বারীদের দৃষ্টি এড়াইয়া জীর্ণদেহ বালক সহিত একটি অতি শীর্ণকায়া রমণী গৃহে প্রবেশ করিয়া ক্ষীণস্বরে কহিল বাবু দুটি খেতে দাও")
output :
| lemma | document |
|---|---|
| [‘একদিন’, ‘প্রাতঃ’, ‘বৈদ্যনাথ’, ‘মার্বলমণ্ডিত’, ‘দালান’, ‘এক’, ‘স্থূলউদর’, ‘সন্ন্যাসী’, ‘দুইসের’, ‘মোহনভোগ’, ‘এবং’, ‘দেড়সের’, ‘দুগ্ধ’, ‘সেবা’, ‘নিযুক্ত’, ‘আছে’, ‘বৈদ্যনাথ’, ‘গা’, ‘একখান’, ‘চাদর’, ‘দেওয়া’, ‘জোড়কর’, ‘একান্ত’, ‘বিনীতভাব’, ‘ভূতল’, ‘বসা’, ‘ভক্তিভরা’, ‘পবিত্র’, ‘ভোজনব্যাপার’, ‘নিরীক্ষণ’, ‘করা’, ‘এমন’, ‘সময়’, ‘কোনোমত’, ‘দ্বারী’, ‘দৃষ্টি’, ‘এড়ানো’, ‘জীর্ণদেহ’, ‘বালক’, ‘সহিত’, ‘এক’, ‘অতি’, ‘শীর্ণকায়া’, ‘রমণী’, ‘গৃহ’, ‘প্রবেশ’, ‘বিশ্বাস’, ‘ক্ষীণস্বর’, ‘কহা’, ‘বাবু’, ‘দুই’, ‘খাওয়া’, ‘দাওয়া’] | একদিন প্রাতে বৈদ্যনাথের মার্বলমণ্ডিত দালানে একটি স্থূলোদর সন্ন্যাসী দুইসের মোহনভোগ এবং দেড়সের দুগ্ধ সেবায় নিযুক্ত আছে বৈদ্যনাথ গায়ে একখানি চাদর দিয়া জোড়করে একান্ত বিনীতভাবে ভূতলে বসিয়া ভক্তিভরে পবিত্র ভোজনব্যাপার নিরীক্ষণ করিতেছিলেন এমন সময় কোনোমতে দ্বারীদের দৃষ্টি এড়াইয়া জীর্ণদেহ বালক সহিত একটি অতি শীর্ণকায়া রমণী গৃহে প্রবেশ করিয়া ক্ষীণস্বরে কহিল বাবু দুটি খেতে দাও |
Japanese Lemmatizer
#Japanese for : Some residents were uncomfortable with this, but it seems that no one is now openly protesting or protesting.
nlu.load("ja.lemma").predict("これに不快感を示す住民はいましたが,現在,表立って反対や抗議の声を挙げている住民はいないようです。")
output :
| lemma | document |
|---|---|
| [‘これ’, ‘にる’, ‘不快’, ‘感’, ‘を’, ‘示す’, ‘住民’, ‘はる’, ‘いる’, ‘まする’, ‘たる’, ‘がる’, ‘,’, ‘現在’, ‘,’, ‘表立つ’, ‘てる’, ‘反対’, ‘やる’, ‘抗議’, ‘のる’, ‘声’, ‘を’, ‘挙げる’, ‘てる’, ‘いる’, ‘住民’, ‘はる’, ‘いる’, ‘なぐ’, ‘よう’, ‘です’, ‘。’] | これに不快感を示す住民はいましたが,現在,表立って反対や抗議の声を挙げている住民はいないようです。 |
Amharic Lemmatizer
#Aharic for : Bookmark the permalink.
nlu.load("am.lemma").predict("መጽሐፉን መጽሐፍ ኡ ን አስያዛት አስያዝ ኧ ኣት ።")
output :
| lemma | document |
|---|---|
| [‘’, ‘መጽሐፍ’, ‘ኡ’, ‘ን’, ‘’, ‘አስያዝ’, ‘ኧ’, ‘ኣት’, ‘።’] | መጽሐፉን መጽሐፍ ኡ ን አስያዛት አስያዝ ኧ ኣት ። |
Bhojpuri Lemmatizer
#Bhojpuri for : In this event, participation of World Bhojpuri Conference, Purvanchal Ekta Manch, Veer Kunwar Singh Foundation, Purvanchal Bhojpuri Mahasabha, and Herf - Media.
nlu.load("bh.lemma").predict("एह आयोजन में विश्व भोजपुरी सम्मेलन , पूर्वांचल एकता मंच , वीर कुँवर सिंह फाउन्डेशन , पूर्वांचल भोजपुरी महासभा , अउर हर्फ - मीडिया के सहभागिता बा ।")
output :
| lemma | document |
|---|---|
| [‘एह’, ‘आयोजन’, ‘में’, ‘विश्व’, ‘भोजपुरी’, ‘सम्मेलन’, ‘COMMA’, ‘पूर्वांचल’, ‘एकता’, ‘मंच’, ‘COMMA’, ‘वीर’, ‘कुँवर’, ‘सिंह’, ‘फाउन्डेशन’, ‘COMMA’, ‘पूर्वांचल’, ‘भोजपुरी’, ‘महासभा’, ‘COMMA’, ‘अउर’, ‘हर्फ’, ‘-‘, ‘मीडिया’, ‘को’, ‘सहभागिता’, ‘बा’, ‘।’] | एह आयोजन में विश्व भोजपुरी सम्मेलन , पूर्वांचल एकता मंच , वीर कुँवर सिंह फाउन्डेशन , पूर्वांचल भोजपुरी महासभा , अउर हर्फ - मीडिया के सहभागिता बा । |
Named Entity Recognition - BERT Tiny (OntoNotes)
nlu.load("en.ner.onto.bert.small_l2_128").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.8536999821662903, 0.7195000052452087, 0.746…] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘1970s’, ‘1980s’, ‘Seattle’, ‘Washington’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] |
Named Entity Recognition - BERT Mini (OntoNotes)
nlu.load("en.ner.onto.bert.small_l4_256").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.835099995136261, 0.40450000762939453, 0.331…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘1970s and 1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘ORG’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘GPE’, ‘GPE’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - BERT Small (OntoNotes)
nlu.load("en.ner.onto.bert.small_l4_512").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.964900016784668, 0.8299000263214111, 0.9607…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘the 1970s and 1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - BERT Medium (OntoNotes)
nlu.load("en.ner.onto.bert.small_l8_512").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.916700005531311, 0.5873000025749207, 0.8816…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘the 1970s and 1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - BERT Base (OntoNotes)
nlu.load("en.ner.onto.bert.cased_base").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.504800021648407, 0.47290000319480896, 0.462…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘the 1970s and 1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - BERT Large (OntoNotes)
nlu.load("en.ner.onto.electra.uncased_small").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.7213000059127808, 0.6384000182151794, 0.731…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘1970s’, ‘1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - ELECTRA Small (OntoNotes)
nlu.load("en.ner.onto.electra.uncased_small").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadella.""",output_level = "document")
output :
| ner_confidence | Entities_classes | entities |
|---|---|---|
| [0.8496000170707703, 0.4465999901294708, 0.568…] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘1970s’, ‘1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] |
Named Entity Recognition - ELECTRA Base (OntoNotes)
nlu.load("en.ner.onto.electra.uncased_base").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadellabase.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.5134000182151794, 0.9419000148773193, 0.802…] | [‘William Henry Gates III’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘the 1970s’, ‘1980s’, ‘Seattle’, ‘Washington’, ‘Gates’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’] |
Named Entity Recognition - ELECTRA Large (OntoNotes)
nlu.load("en.ner.onto.electra.uncased_large").predict("""William Henry Gates III (born October 28, 1955) is an American business magnate,
software developer, investor, and philanthropist. He is best known as the co-founder of Microsoft Corporation. During his career at Microsoft,
Gates held the positions of chairman, chief executive officer (CEO), president and chief software architect,
while also being the largest individual shareholder until May 2014.
He is one of the best-known entrepreneurs and pioneers of the microcomputer revolution of the 1970s and 1980s. Born and raised in Seattle, Washington, Gates co-founded Microsoft with childhood friend Paul Allen in 1975, in Albuquerque, New Mexico;
it went on to become the world's largest personal computer software company. Gates led the company as chairman and CEO until stepping down as CEO in January 2000, but he remained chairman and became chief software architect.
During the late 1990s, Gates had been criticized for his business tactics, which have been considered anti-competitive. This opinion has been upheld by numerous court rulings. In June 2006, Gates announced that he would be transitioning to a part-time
role at Microsoft and full-time work at the Bill & Melinda Gates Foundation, the private charitable foundation that he and his wife, Melinda Gates, established in 2000.
He gradually transferred his duties to Ray Ozzie and Craig Mundie.
He stepped down as chairman of Microsoft in February 2014 and assumed a new post as technology adviser to support the newly appointed CEO Satya Nadellabase.""",output_level = "document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.8442000150680542, 0.26840001344680786, 0.57…] | [‘William Henry Gates’, ‘October 28, 1955’, ‘American’, ‘Microsoft Corporation’, ‘Microsoft’, ‘Gates’, ‘May 2014’, ‘one’, ‘1970s’, ‘1980s’, ‘Seattle’, ‘Washington’, ‘Gates co-founded’, ‘Microsoft’, ‘Paul Allen’, ‘1975’, ‘Albuquerque’, ‘New Mexico’, ‘largest’] | [‘PERSON’, ‘DATE’, ‘NORP’, ‘ORG’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘CARDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘GPE’, ‘PERSON’, ‘ORG’, ‘PERSON’, ‘DATE’, ‘GPE’, ‘GPE’, ‘GPE’] |
Recognize Entities OntoNotes - BERT Tiny
nlu.load("en.ner.onto.bert.tiny").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.994700014591217, 0.9412999749183655, 0.9685…] | [‘Johnson’, ‘first’, ‘2001’, ‘Parliament’, ‘eight years’, ‘London’, ‘2008 to 2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘ORG’, ‘DATE’, ‘GPE’, ‘DATE’] |
Recognize Entities OntoNotes - BERT Mini
nlu.load("en.ner.onto.bert.mini").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.996399998664856, 0.9733999967575073, 0.8766…] | [‘Johnson’, ‘first’, ‘2001’, ‘eight years’, ‘London’, ‘2008 to 2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘DATE’] |
Recognize Entities OntoNotes - BERT Small
nlu.load("en.ner.onto.bert.small").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9987999796867371, 0.9610000252723694, 0.998…] | [‘Johnson’, ‘first’, ‘2001’, ‘eight years’, ‘London’, ‘2008 to 2016’, ‘Parliament’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘DATE’, ‘ORG’] |
Recognize Entities OntoNotes - BERT Medium
nlu.load("en.ner.onto.bert.medium").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9969000220298767, 0.8575999736785889, 0.995…] | [‘Johnson’, ‘first’, ‘2001’, ‘eight years’, ‘London’, ‘2008 to 2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘DATE’] |
Recognize Entities OntoNotes - BERT Base
nlu.load("en.ner.onto.bert.base").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.996999979019165, 0.933899998664856, 0.99930…] | [‘Johnson’, ‘first’, ‘2001’, ‘Parliament’, ‘eight years’, ‘London’, ‘2008 to 2016’, ‘Parliament’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘ORG’, ‘DATE’, ‘GPE’, ‘DATE’, ‘ORG’] |
Recognize Entities OntoNotes - BERT Large
nlu.load("en.ner.onto.bert.large").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9786999821662903, 0.9549000263214111, 0.998…] | [‘Johnson’, ‘first’, ‘2001’, ‘Parliament’, ‘eight years’, ‘London’, ‘2008 to 2016’, ‘Parliament’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘ORG’, ‘DATE’, ‘GPE’, ‘DATE’, ‘ORG’] |
Recognize Entities OntoNotes - ELECTRA Small
nlu.load("en.ner.onto.electra.small").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9952999949455261, 0.8589000105857849, 0.996…] | [‘Johnson’, ‘first’, ‘2001’, ‘eight years’, ‘London’, ‘2008 to 2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘DATE’] |
Recognize Entities OntoNotes - ELECTRA Base
nlu.load("en.ner.onto.electra.base").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9987999796867371, 0.9474999904632568, 0.999…] | [‘Johnson’, ‘first’, ‘2001’, ‘Parliament’, ‘eight years’, ‘London’, ‘2008’, ‘2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘ORG’, ‘DATE’, ‘GPE’, ‘DATE’, ‘DATE’] |
Recognize Entities OntoNotes - ELECTRA Large
nlu.load("en.ner.onto.large").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London, from 2008 to 2016, before rejoining Parliament.",output_level="document")
output :
| ner_confidence | entities | Entities_classes |
|---|---|---|
| [0.9998000264167786, 0.9613999724388123, 0.998…] | [‘Johnson’, ‘first’, ‘2001’, ‘eight years’, ‘London’, ‘2008 to 2016’] | [‘PERSON’, ‘ORDINAL’, ‘DATE’, ‘DATE’, ‘GPE’, ‘DATE’] |
NLU Installation
# PyPi
!pip install nlu pyspark==2.4.7
#Conda
# Install NLU from Anaconda/Conda
conda install -os_components johnsnowlabs nlu
Additional NLU ressources
NLU Version 1.1.1
We are very excited to release NLU 1.1.1! This release features 3 new tutorial notebooks for Open/Closed book question answering with Google’s T5, Intent classification and Aspect Based NER. In Addition NLU 1.1.0 comes with 25+ pretrained models and pipelines in Amharic, Bengali, Bhojpuri, Japanese, and Korean languages from the amazing Spark2.7.2 release Finally NLU now supports running on Spark 2.3 clusters.
NLU 1.1.1 New Non-English Models
| Language | nlu.load() reference | Spark NLP Model reference | Type |
|---|---|---|---|
| Arabic | ar.ner | arabic_w2v_cc_300d | Named Entity Recognizer |
| Arabic | ar.embed.aner | aner_cc_300d | Word Embedding |
| Arabic | ar.embed.aner.300d | aner_cc_300d | Word Embedding (Alias) |
| Bengali | bn.stopwords | stopwords_bn | Stopwords Cleaner |
| Bengali | bn.pos | pos_msri | Part of Speech |
| Thai | th.segment_words | wordseg_best | Word Segmenter |
| Thai | th.pos | pos_lst20 | Part of Speech |
| Thai | th.sentiment | sentiment_jager_use | Sentiment Classifier |
| Thai | th.classify.sentiment | sentiment_jager_use | Sentiment Classifier (Alias) |
| Chinese | zh.pos.ud_gsd_trad | pos_ud_gsd_trad | Part of Speech |
| Chinese | zh.segment_words.gsd | wordseg_gsd_ud_trad | Word Segmenter |
| Bihari | bh.pos | pos_ud_bhtb | Part of Speech |
| Amharic | am.pos | pos_ud_att | Part of Speech |
NLU 1.1.1 New English Models and Pipelines
New Easy NLU 1-liner Examples :
Extract aspects and entities from airline questions (ATIS dataset)
nlu.load("en.ner.atis").predict("i want to fly from baltimore to dallas round trip")
output: ["baltimore"," dallas", "round trip"]
Intent Classification for Airline Traffic Information System queries (ATIS dataset)
nlu.load("en.classify.questions.atis").predict("what is the price of flight from newyork to washington")
output: "atis_airfare"
Recognize Entities OntoNotes - ELECTRA Large
nlu.load("en.ner.onto.large").predict("Johnson first entered politics when elected in 2001 as a member of Parliament. He then served eight years as the mayor of London.")
output: ["Johnson", "first", "2001", "eight years", "London"]
Question classification of open-domain and fact-based questions Pipeline - TREC50
nlu.load("en.classify.trec50.component_list").predict("When did the construction of stone circles begin in the UK? ")
output: LOC_other
Traditional Chinese Word Segmentation
# 'However, this treatment also creates some problems' in Chinese
nlu.load("zh.segment_words.gsd").predict("然而,這樣的處理也衍生了一些問題。")
output: ["然而",",","這樣","的","處理","也","衍生","了","一些","問題","。"]
Part of Speech for Traditional Chinese
# 'However, this treatment also creates some problems' in Chinese
nlu.load("zh.pos.ud_gsd_trad").predict("然而,這樣的處理也衍生了一些問題。")
Output:
| Token | POS |
|---|---|
| 然而 | ADV |
| , | PUNCT |
| 這樣 | PRON |
| 的 | PART |
| 處理 | NOUN |
| 也 | ADV |
| 衍生 | VERB |
| 了 | PART |
| 一些 | ADJ |
| 問題 | NOUN |
| 。 | PUNCT |
Thai Word Segment Recognition
# 'Mona Lisa is a 16th-century oil painting created by Leonardo held at the Louvre in Paris' in Thai
nlu.loadnlu.load("th.segment_words").predict("Mona Lisa เป็นภาพวาดสีน้ำมันในศตวรรษที่ 16 ที่สร้างโดย Leonardo จัดขึ้นที่พิพิธภัณฑ์ลูฟร์ในปารีส")
Output:
| token |
|---|
| M |
| o |
| n |
| a |
| Lisa |
| เป็น |
| ภาพ |
| ว |
| า |
| ด |
| สีน้ำ |
| มัน |
| ใน |
| ศตวรรษ |
| ที่ |
| 16 |
| ที่ |
| สร้าง |
| โ |
| ด |
| ย |
| L |
| e |
| o |
| n |
| a |
| r |
| d |
| o |
| จัด |
| ขึ้น |
| ที่ |
| พิพิธภัณฑ์ |
| ลูฟร์ |
| ใน |
| ปารีส |
Part of Speech for Bengali (POS)
# 'The village is also called 'Mod' in Tora language' in Behgali
nlu.load("bn.pos").predict("বাসস্থান-ঘরগৃহস্থালি তোড়া ভাষায় গ্রামকেও বলে ` মোদ ' ৷")
Output:
| token | pos |
|---|---|
| বাসস্থান-ঘরগৃহস্থালি | NN |
| তোড়া | NNP |
| ভাষায় | NN |
| গ্রামকেও | NN |
| বলে | VM |
| ` | SYM |
| মোদ | NN |
| ’ | SYM |
| ৷ | SYM |
Stop Words Cleaner for Bengali
# 'This language is not enough' in Bengali
df = nlu.load("bn.stopwords").predict("এই ভাষা যথেষ্ট নয়")
Output:
| cleanTokens | token |
|---|---|
| ভাষা | এই |
| যথেষ্ট | ভাষা |
| নয় | যথেষ্ট |
| None | নয় |
Part of Speech for Bengali
# 'The people of Ohu know that the foundation of Bhojpuri was shaken' in Bengali
nlu.load('bh.pos').predict("ओहु लोग के मालूम बा कि श्लील होखते भोजपुरी के नींव हिल जाई")
Output:
| pos | token |
|---|---|
| DET | ओहु |
| NOUN | लोग |
| ADP | के |
| NOUN | मालूम |
| VERB | बा |
| SCONJ | कि |
| ADJ | श्लील |
| VERB | होखते |
| PROPN | भोजपुरी |
| ADP | के |
| NOUN | नींव |
| VERB | हिल |
| AUX | जाई |
Amharic Part of Speech (POS)
# ' "Son, finish the job," he said.' in Amharic
nlu.load('am.pos').predict('ልጅ ኡ ን ሥራ ው ን አስጨርስ ኧው ኣል ኧሁ ።"')
Output:
| pos | token |
|---|---|
| NOUN | ልጅ |
| DET | ኡ |
| PART | ን |
| NOUN | ሥራ |
| DET | ው |
| PART | ን |
| VERB | አስጨርስ |
| PRON | ኧው |
| AUX | ኣል |
| PRON | ኧሁ |
| PUNCT | ። |
| NOUN | ” |
Thai Sentiment Classification
# 'I love peanut butter and jelly!' in thai
nlu.load('th.classify.sentiment').predict('ฉันชอบเนยถั่วและเยลลี่!')[['sentiment','sentiment_confidence']]
Output:
| sentiment | sentiment_confidence |
|---|---|
| positive | 0.999998 |
Arabic Named Entity Recognition (NER)
# 'In 1918, the forces of the Arab Revolt liberated Damascus with the help of the British' in Arabic
nlu.load('ar.ner').predict('في عام 1918 حررت قوات الثورة العربية دمشق بمساعدة من الإنكليز',output_level='chunk')[['entities_confidence','ner_confidence','entities']]
Output:
| entity_class | ner_confidence | entities |
|---|---|---|
| ORG | [1.0, 1.0, 1.0, 0.9997000098228455, 0.9840999841690063, 0.9987999796867371, 0.9990000128746033, 0.9998999834060669, 0.9998999834060669, 0.9993000030517578, 0.9998999834060669] | قوات الثورة العربية |
| LOC | [1.0, 1.0, 1.0, 0.9997000098228455, 0.9840999841690063, 0.9987999796867371, 0.9990000128746033, 0.9998999834060669, 0.9998999834060669, 0.9993000030517578, 0.9998999834060669] | دمشق |
| PER | [1.0, 1.0, 1.0, 0.9997000098228455, 0.9840999841690063, 0.9987999796867371, 0.9990000128746033, 0.9998999834060669, 0.9998999834060669, 0.9993000030517578, 0.9998999834060669] | الإنكليز |
NLU 1.1.1 Enhancements :
- Spark 2.3 compatibility
New NLU Notebooks and Tutorials
- Open and Closed book question Ansering
- Aspect based NER for Airline ATIS
- Intent Classification for Airline emssages ATIS
Installation
# PyPi
!pip install nlu pyspark==2.4.7
#Conda
# Install NLU from Anaconda/Conda
conda install -os_components johnsnowlabs nlu
Additional NLU ressources
NLU Version 1.1.0
We are incredibly excited to release NLU 1.1.0!
This release integrates the 720+ new models from the latest Spark-NLP 2.7.0 + releases.
You can now achieve state-of-the-art results with Sequence2Sequence transformers for problems like text summarization, question answering, translation between 192+ languages and extract Named Entity in various Right to Left written languages like Korean, Japanese, Chinese and many more in 1 line of code!
These new features are possible because of the integration of the Google’s T5 models and Microsoft’s Marian models transformers
NLU 1.1.0 has over 720+ new pretrained models and pipelines while extending the support of multi-lingual models to 192+ languages such as Chinese, Japanese, Korean, Arabic, Persian, Urdu, and Hebrew.
NLU 1.1.0 New Features
- 720+ new models you can find an overview of all NLU models here and further documentation in the models hub
- NEW: Introducing MarianTransformer annotator for machine translation based on MarianNMT models. Marian is an efficient, free Neural Machine Translation framework mainly being developed by the Microsoft Translator team (646+ pretrained models & pipelines in 192+ languages)
- NEW: Introducing T5Transformer annotator for Text-To-Text Transfer Transformer (Google T5) models to achieve state-of-the-art results on multiple NLP tasks such as Translation, Summarization, Question Answering, Sentence Similarity, and so on
- NEW: Introducing brand new and refactored language detection and identification models. The new LanguageDetectorDL is faster, more accurate, and supports up to 375 languages
- NEW: Introducing WordSegmenter model for word segmentation of languages without any rule-based tokenization such as Chinese, Japanese, or Korean
- NEW: Introducing DocumentNormalizer component for cleaning content from HTML or XML documents, applying either data cleansing using an arbitrary number of custom regular expressions either data extraction following the different parameters
NLU 1.1.0 New Notebooks, Tutorials and Articles
- Translate between 192+ languages with marian
- Try out the 18 Tasks like Summarization Question Answering and more on T5
- Tokenize, extract POS and NER in Chinese
- Tokenize, extract POS and NER in Korean
- Tokenize, extract POS and NER in Japanese
- Normalize documents
- Aspect based sentiment NER sentiment for restaurants
NLU 1.1.0 New Training Tutorials
Binary Classifier training Jupyter tutorials
Multi Class text Classifier training Jupyter tutorials
NLU 1.1.0 New Medium Tutorials
- 1 line to Glove Word Embeddings with NLU with t-SNE plots
- 1 line to Xlnet Word Embeddings with NLU with t-SNE plots
- 1 line to AlBERT Word Embeddings with NLU with t-SNE plots
- 1 line to CovidBERT Word Embeddings with NLU with t-SNE plots
- 1 line to Electra Word Embeddings with NLU with t-SNE plots
- 1 line to BioBERT Word Embeddings with NLU with t-SNE plots
Translation
Translation example
You can translate between more than 192 Languages pairs with the Marian Models
You need to specify the language your data is in as start_language and the language you want to translate to as target_language.
The language references must be ISO language codes
nlu.load('<start_language>.translate.<target_language>')
Translate Turkish to English:
nlu.load('tr.translate_to.fr')
Translate English to French:
nlu.load('en.translate_to.fr')
Translate French to Hebrew:
nlu.load('en.translate_to.fr')
translate_pipe = nlu.load('en.translate_to.fr')
df = translate_pipe.predict('Billy likes to go to the mall every sunday')
df
| sentence | translation |
|---|---|
| Billy likes to go to the mall every sunday | Billy geht gerne jeden Sonntag ins Einkaufszentrum |
Overview of every task available with T5
The T5 model is trained on various datasets for 17 different tasks which fall into 8 categories.
- Text summarization
- Question answering
- Translation
- Sentiment analysis
- Natural Language inference
- Coreference resolution
- Sentence Completion
- Word sense disambiguation
Every T5 Task with explanation:
| Task Name | Explanation |
|---|---|
| 1.CoLA | Classify if a sentence is gramaticaly correct |
| 2.RTE | Classify whether if a statement can be deducted from a sentence |
| 3.MNLI | Classify for a hypothesis and premise whether they contradict or contradict each other or neither of both (3 class). |
| 4.MRPC | Classify whether a pair of sentences is a re-phrasing of each other (semantically equivalent) |
| 5.QNLI | Classify whether the answer to a question can be deducted from an answer candidate. |
| 6.QQP | Classify whether a pair of questions is a re-phrasing of each other (semantically equivalent) |
| 7.SST2 | Classify the sentiment of a sentence as positive or negative |
| 8.STSB | Classify the sentiment of a sentence on a scale from 1 to 5 (21 Sentiment classes) |
| 9.CB | Classify for a premise and a hypothesis whether they contradict each other or not (binary). |
| 10.COPA | Classify for a question, premise, and 2 choices which choice the correct choice is (binary). |
| 11.MultiRc | Classify for a question, a paragraph of text, and an answer candidate, if the answer is correct (binary), |
| 12.WiC | Classify for a pair of sentences and a disambigous word if the word has the same meaning in both sentences. |
| 13.WSC/DPR | Predict for an ambiguous pronoun in a sentence what it is referring to. |
| 14.Summarization | Summarize text into a shorter representation. |
| 15.SQuAD | Answer a question for a given context. |
| 16.WMT1. | Translate English to German |
| 17.WMT2. | Translate English to French |
| 18.WMT3. | Translate English to Romanian |
refer to this notebook to see how to use every T5 Task.
Question Answering
Predict an answer to a question based on input context.
This is based on SQuAD - Context based question answering
| Predicted Answer | Question | Context |
|---|---|---|
| carbon monoxide | What does increased oxygen concentrations in the patient’s lungs displace? | Hyperbaric (high-pressure) medicine uses special oxygen chambers to increase the partial pressure of O 2 around the patient and, when needed, the medical staff. Carbon monoxide poisoning, gas gangrene, and decompression sickness (the ’bends’) are sometimes treated using these devices. Increased O 2 concentration in the lungs helps to displace carbon monoxide from the heme group of hemoglobin. Oxygen gas is poisonous to the anaerobic bacteria that cause gas gangrene, so increasing its partial pressure helps kill them. Decompression sickness occurs in divers who decompress too quickly after a dive, resulting in bubbles of inert gas, mostly nitrogen and helium, forming in their blood. Increasing the pressure of O 2 as soon as possible is part of the treatment. |
| pie | What did Joey eat for breakfast? | Once upon a time, there was a squirrel named Joey. Joey loved to go outside and play with his cousin Jimmy. Joey and Jimmy played silly games together, and were always laughing. One day, Joey and Jimmy went swimming together 50 at their Aunt Julie’s pond. Joey woke up early in the morning to eat some food before they left. Usually, Joey would eat cereal, fruit (a pear), or oatmeal for breakfast. After he ate, he and Jimmy went to the pond. On their way there they saw their friend Jack Rabbit. They dove into the water and swam for several hours. The sun was out, but the breeze was cold. Joey and Jimmy got out of the water and started walking home. Their fur was wet, and the breeze chilled them. When they got home, they dried off, and Jimmy put on his favorite purple shirt. Joey put on a blue shirt with red and green dots. The two squirrels ate some food that Joey’s mom, Jasmine, made and went off to bed,’ |
# Set the task on T5
t5['t5'].setTask('question ')
# define Data, add additional tags between sentences
data = ['''
What does increased oxygen concentrations in the patient’s lungs displace?
context: Hyperbaric (high-pressure) medicine uses special oxygen chambers to increase the partial pressure of O 2 around the patient and, when needed, the medical staff. Carbon monoxide poisoning, gas gangrene, and decompression sickness (the ’bends’) are sometimes treated using these devices. Increased O 2 concentration in the lungs helps to displace carbon monoxide from the heme group of hemoglobin. Oxygen gas is poisonous to the anaerobic bacteria that cause gas gangrene, so increasing its partial pressure helps kill them. Decompression sickness occurs in divers who decompress too quickly after a dive, resulting in bubbles of inert gas, mostly nitrogen and helium, forming in their blood. Increasing the pressure of O 2 as soon as possible is part of the treatment.
''']
#Predict on text data with T5
t5.predict(data)
How to configure T5 task parameter for Squad Context based question answering and pre-process data
.setTask('question:) and prefix the context which can be made up of multiple sentences with context:
Example pre-processed input for T5 Squad Context based question answering:
question: What does increased oxygen concentrations in the patient’s lungs displace?
context: Hyperbaric (high-pressure) medicine uses special oxygen chambers to increase the partial pressure of O 2 around the patient and, when needed, the medical staff. Carbon monoxide poisoning, gas gangrene, and decompression sickness (the ’bends’) are sometimes treated using these devices. Increased O 2 concentration in the lungs helps to displace carbon monoxide from the heme group of hemoglobin. Oxygen gas is poisonous to the anaerobic bacteria that cause gas gangrene, so increasing its partial pressure helps kill them. Decompression sickness occurs in divers who decompress too quickly after a dive, resulting in bubbles of inert gas, mostly nitrogen and helium, forming in their blood. Increasing the pressure of O 2 as soon as possible is part of the treatment.
Text Summarization
Summarizes a paragraph into a shorter version with the same semantic meaning, based on Text summarization
# Set the task on T5
pipe = nlu.load('summarize')
# define Data, add additional tags between sentences
data = [
'''
The belgian duo took to the dance floor on monday night with some friends . manchester united face newcastle in the premier league on wednesday . red devils will be looking for just their second league away win in seven . louis van gaal’s side currently sit two points clear of liverpool in fourth .
''',
''' Calculus, originally called infinitesimal calculus or "the calculus of infinitesimals", is the mathematical study of continuous change, in the same way that geometry is the study of shape and algebra is the study of generalizations of arithmetic operations. It has two major branches, differential calculus and integral calculus; the former concerns instantaneous rates of change, and the slopes of curves, while integral calculus concerns accumulation of quantities, and areas under or between curves. These two branches are related to each other by the fundamental theorem of calculus, and they make use of the fundamental notions of convergence of infinite sequences and infinite series to a well-defined limit.[1] Infinitesimal calculus was developed independently in the late 17th century by Isaac Newton and Gottfried Wilhelm Leibniz.[2][3] Today, calculus has widespread uses in science, engineering, and economics.[4] In mathematics education, calculus denotes courses of elementary mathematical analysis, which are mainly devoted to the study of functions and limits. The word calculus (plural calculi) is a Latin word, meaning originally "small pebble" (this meaning is kept in medicine – see Calculus (medicine)). Because such pebbles were used for calculation, the meaning of the word has evolved and today usually means a method of computation. It is therefore used for naming specific methods of calculation and related theories, such as propositional calculus, Ricci calculus, calculus of variations, lambda calculus, and process calculus.'''
]
#Predict on text data with T5
pipe.predict(data)
| Predicted summary | Text |
|---|---|
| manchester united face newcastle in the premier league on wednesday . louis van gaal’s side currently sit two points clear of liverpool in fourth . the belgian duo took to the dance floor on monday night with some friends . | the belgian duo took to the dance floor on monday night with some friends . manchester united face newcastle in the premier league on wednesday . red devils will be looking for just their second league away win in seven . louis van gaal’s side currently sit two points clear of liverpool in fourth . |
Binary Sentence similarity/ Paraphrasing
Binary sentence similarity example
Classify whether one sentence is a re-phrasing or similar to another sentence
This is a sub-task of GLUE and based on MRPC - Binary Paraphrasing/ sentence similarity classification
t5 = nlu.load('en.t5.base')
# Set the task on T5
t5['t5'].setTask('mrpc ')
# define Data, add additional tags between sentences
data = [
''' sentence1: We acted because we saw the existing evidence in a new light , through the prism of our experience on 11 September , " Rumsfeld said .
sentence2: Rather , the US acted because the administration saw " existing evidence in a new light , through the prism of our experience on September 11 "
'''
,
'''
sentence1: I like to eat peanutbutter for breakfast
sentence2: I like to play football.
'''
]
#Predict on text data with T5
t5.predict(data)
| Sentence1 | Sentence2 | prediction |
|---|---|---|
| We acted because we saw the existing evidence in a new light , through the prism of our experience on 11 September , “ Rumsfeld said . | Rather , the US acted because the administration saw “ existing evidence in a new light , through the prism of our experience on September 11 “ . | equivalent |
| I like to eat peanutbutter for breakfast | I like to play football | not_equivalent |
How to configure T5 task for MRPC and pre-process text
.setTask('mrpc sentence1:) and prefix second sentence with sentence2:
Example pre-processed input for T5 MRPC - Binary Paraphrasing/ sentence similarity
mrpc
sentence1: We acted because we saw the existing evidence in a new light , through the prism of our experience on 11 September , " Rumsfeld said .
sentence2: Rather , the US acted because the administration saw " existing evidence in a new light , through the prism of our experience on September 11",
Regressive Sentence similarity/ Paraphrasing
Measures how similar two sentences are on a scale from 0 to 5 with 21 classes representing a regressive label.
This is a sub-task of GLUE and based onSTSB - Regressive semantic sentence similarity .
t5 = nlu.load('en.t5.base')
# Set the task on T5
t5['t5'].setTask('stsb ')
# define Data, add additional tags between sentences
data = [
''' sentence1: What attributes would have made you highly desirable in ancient Rome?
sentence2: How I GET OPPERTINUTY TO JOIN IT COMPANY AS A FRESHER?'
'''
,
'''
sentence1: What was it like in Ancient rome?
sentence2: What was Ancient rome like?
''',
'''
sentence1: What was live like as a King in Ancient Rome??
sentence2: What was Ancient rome like?
'''
]
#Predict on text data with T5
t5.predict(data)
| Question1 | Question2 | prediction |
|---|---|---|
| What attributes would have made you highly desirable in ancient Rome? | How I GET OPPERTINUTY TO JOIN IT COMPANY AS A FRESHER? | 0 |
| What was it like in Ancient rome? | What was Ancient rome like? | 5.0 |
| What was live like as a King in Ancient Rome?? | What is it like to live in Rome? | 3.2 |
How to configure T5 task for stsb and pre-process text
.setTask('stsb sentence1:) and prefix second sentence with sentence2:
Example pre-processed input for T5 STSB - Regressive semantic sentence similarity
stsb
sentence1: What attributes would have made you highly desirable in ancient Rome?
sentence2: How I GET OPPERTINUTY TO JOIN IT COMPANY AS A FRESHER?',
Grammar Checking
Grammar checking with T5 example
Judges if a sentence is grammatically acceptable.
Based on CoLA - Binary Grammatical Sentence acceptability classification
pipe = nlu.load('grammar_correctness')
# Set the task on T5
pipe['t5'].setTask('cola sentence: ')
# define Data
data = ['Anna and Mike is going skiing and they is liked is','Anna and Mike like to dance']
#Predict on text data with T5
pipe.predict(data)
| sentence | prediction |
|---|---|
| Anna and Mike is going skiing and they is liked is | unacceptable |
| Anna and Mike like to dance | acceptable |
Document Normalization
Document Normalizer example
The DocumentNormalizer extracts content from HTML or XML documents, applying either data cleansing using an arbitrary number of custom regular expressions either data extraction following the different parameters
pipe = nlu.load('norm_document')
data = '<!DOCTYPE html> <html> <head> <title>Example</title> </head> <body> <p>This is an example of a simple HTML page with one paragraph.</p> </body> </html>'
df = pipe.predict(data,output_level='document')
df
| text | normalized_text |
|---|---|
<!DOCTYPE html> <html> <head> <title>Example</title> </head> <body> <p>This is an example of a simple HTML page with one paragraph.</p> </body> </html> |
Example This is an example of a simple HTML page with one paragraph. |
Word Segmenter
Word Segmenter Example
The WordSegmenter segments languages without any rule-based tokenization such as Chinese, Japanese, or Korean
pipe = nlu.load('ja.segment_words')
# japanese for 'Donald Trump and Angela Merkel dont share many opinions'
ja_data = ['ドナルド・トランプとアンゲラ・メルケルは多くの意見を共有していません']
df = pipe.predict(ja_data, output_level='token')
df
| token |
|---|
| ドナルド |
| ・ |
| トランプ |
| と |
| アンゲラ |
| ・ |
| メルケル |
| は |
| 多く |
| の |
| 意見 |
| を |
| 共有 |
| し |
| て |
| い |
| ませ |
| ん |
Installation
# PyPi
!pip install nlu pyspark==2.4.7
#Conda
# Install NLU from Anaconda/Conda
conda install -os_components johnsnowlabs nlu
Additional NLU ressources
NLU Version 1.0.6
Trainable Multi Label Classifiers, predict Stackoverflow Tags and much more in 1 Line of with NLU 1.0.6
We are glad to announce NLU 1.0.6 has been released! NLU 1.0.6 comes with the Multi Label classifier, it can learn to map strings to multiple labels. The Multi Label Classifier is using Bidirectional GRU and CNNs inside TensorFlow and supports up to 100 classes.
NLU 1.0.6 New Features
- Multi Label Classifier
- The Multi Label Classifier learns a 1 to many mapping between text and labels. This means it can predict multiple labels at the same time for a given input string. This is very helpful for tasks similar to content tag prediction (HashTags/RedditTags/YoutubeTags/Toxic/E2e etc..)
- Support up to 100 classes
- Pre-trained Multi Label Classifiers are already avaiable as Toxic and E2E classifiers
Multi Label Classifier
- Train Multi Label Classifier on E2E dataset Demo
- Train Multi Label Classifier on Stack Overflow Question Tags dataset Demo
This model can predict multiple labels for one sentence. To train the Multi Label text classifier model, you must pass a dataframe with atextcolumn and aycolumn for the label.
Theylabel must be a string column where each label is seperated with a seperator.
By default,,is assumed as line seperator.
If your dataset is using a different label seperator, you must configure thelabel_seperatorparameter while calling thefit()method.
By default Universal Sentence Encoder Embeddings (USE) are used as sentence embeddings for training.
fitted_pipe = nlu.load('train.multi_classifier').fit(train_df)
preds = fitted_pipe.predict(train_df)
If you add a nlu sentence embeddings reference, before the train reference, NLU will use that Sentence embeddings instead of the default USE.
#Train on BERT sentence emebddings
fitted_pipe = nlu.load('embed_sentence.bert train.multi_classifier').fit(train_df)
preds = fitted_pipe.predict(train_df)
Configure a custom line seperator
#Use ; as label seperator
fitted_pipe = nlu.load('embed_sentence.electra train.multi_classifier').fit(train_df, label_seperator=';')
preds = fitted_pipe.predict(train_df)
NLU 1.0.6 Enhancements
- Improved outputs for Toxic and E2E Classifier.
- by default, all predicted classes and their confidences which are above the threshold will be returned inside of a list in the Pandas dataframe
- by configuring meta=True, the confidences for all classes will be returned.
NLU Version 1.0.6
NLU 1.0.6 Bug-fixes
- Fixed a bug that caused
en.ner.dl.bertto be inaccessible - Fixed a bug that caused
pt.ner.largeto be inaccessible - Fixed a bug that caused USE embeddings not properly beeing configured to document level output when using multiple embeddings at the same time
NLU Version 1.0.5
Trainable Part of Speech Tagger (POS), Sentiment Classifier with BERT/USE/ELECTRA sentence embeddings in 1 Line of code! Latest NLU Release 1.0.5
We are glad to announce NLU 1.0.5 has been released!
This release comes with a trainable Sentiment classifier and a Trainable Part of Speech (POS) models!
These Neural Network Architectures achieve the state of the art (SOTA) on most binary Sentiment analysis and Part of Speech Tagging tasks!
You can train the Sentiment Model on any of the 100+ Sentence Embeddings which include BERT, ELECTRA, USE, Multi Lingual BERT Sentence Embeddings and many more!
Leverage this and achieve the state of the art in any of your datasets, all of this in just 1 line of Python code
NLU 1.0.5 New Features
- Trainable Sentiment DL classifier
- Trainable POS
NLU 1.0.5 New Notebooks and Tutorials
Sentiment Classifier Training
Sentiment Classification Training Demo
To train the Binary Sentiment classifier model, you must pass a dataframe with a ‘text’ column and a ‘y’ column for the label.
By default Universal Sentence Encoder Embeddings (USE) are used as sentence embeddings.
fitted_pipe = nlu.load('train.sentiment').fit(train_df)
preds = fitted_pipe.predict(train_df)
If you add a nlu sentence embeddings reference, before the train reference, NLU will use that Sentence embeddings instead of the default USE.
#Train Classifier on BERT sentence embeddings
fitted_pipe = nlu.load('embed_sentence.bert train.classifier').fit(train_df)
preds = fitted_pipe.predict(train_df)
#Train Classifier on ELECTRA sentence embeddings
fitted_pipe = nlu.load('embed_sentence.electra train.classifier').fit(train_df)
preds = fitted_pipe.predict(train_df)
Part Of Speech Tagger Training
Part Of Speech Tagger Training demo
fitted_pipe = nlu.load('train.pos').fit(train_df)
preds = fitted_pipe.predict(train_df)
NLU 1.0.5 Installation changes
Starting from version 1.0.5 NLU will not automatically install pyspark for users anymore.
This enables easier customizing the Pyspark version which makes it easier to use in various cluster enviroments.
To install NLU from now on, please run
pip install nlu pyspark==2.4.7
or install any pyspark>=2.4.0 with pyspark<3
NLU 1.0.5 Improvements
- Improved Databricks path handling for loading and storing models.
NLU Version 1.0.4
John Snow Labs NLU 1.0.4 : Trainable Named Entity Recognizer (NER) , achieve SOTA in 1 line of code and easy scaling to 100’s of Spark nodes
We are glad to announce NLU 1.0.4 releases the State of the Art breaking Neural Network architecture for NER, Char CNNs - BiLSTM - CRF!
#fit and predict in 1 line!
nlu.load('train.ner').fit(dataset).predict(dataset)
#fit and predict in 1 line with BERT!
nlu.load('bert train.ner').fit(dataset).predict(dataset)
#fit and predict in 1 line with ALBERT!
nlu.load('albert train.ner').fit(dataset).predict(dataset)
#fit and predict in 1 line with ELMO!
nlu.load('elmo train.ner').fit(dataset).predict(dataset)
Any NLU pipeline stored can now be loaded as pyspark ML pipeline
# Ready for big Data with Spark distributed computing
import pyspark
nlu_pipe.save(path)
pyspark_pipe = pyspark.ml.PipelineModel.load(stored_model_path)
pyspark_pipe.transform(spark_df)
NLU 1.0.4 New Features
- Trainable Named Entity Recognizer
- NLU pipeline loadable as Spark pipelines
NLU 1.0.4 New Notebooks,Tutorials and Docs
- NER training demo
- Multi Class Text Classifier Training Demo updated to showcase usage of different Embeddings
- New Documentation Page on how to train Models with NLU
- Databricks Notebook showcasing Scaling with NLU
NLU 1.0.4 Bug Fixes
- Fixed a bug that NER token confidences do not appear. They now appear when nlu.load(‘ner’).predict(df, meta=True) is called.
- Fixed a bug that caused some Spark NLP models to not be loaded properly in offline mode
NLU Version 1.0.3
We are happy to announce NLU 1.0.3 comes with a lot new features, training classifiers, saving them and loading them offline, enabling running NLU with no internet connection, new notebooks and articles!
NLU 1.0.3 New Features
- Train a Deep Learning classifier in 1 line! The popular ClassifierDL which can achieve state of the art results on any multi class text classification problem is now trainable! All it takes is just nlu.load(‘train.classifier).fit(dataset) . Your dataset can be a Pandas/Spark/Modin/Ray/Dask dataframe and needs to have a column named x for text data and a column named y for labels
- Saving pipelines to HDD is now possible with nlu.save(path)
- Loading pipelines from disk now possible with nlu.load(path=path).
- NLU offline mode: Loading from disk makes running NLU offline now possible, since you can load pipelines/models from your local hard drive instead of John Snow Labs AWS servers.
NLU 1.0.3 New Notebooks and Tutorials
- New colab notebook showcasing nlu training, saving and loading from disk
- Sentence Similarity with BERT, Electra and Universal Sentence Encoder Medium Tutorial
- Sentence Similarity with BERT, Electra and Universal Sentence Encoder
- Train a Deep Learning Classifier
- Sentence Detector Notebook Updated
- New Workshop video
NLU 1.0.3 Bug fixes
- Sentence Detector bugfix
NLU Version 1.0.2
We are glad to announce nlu 1.0.2 is released!
NLU 1.0.2 Enhancements
- More semantically concise output levels sentence and document enforced :
- If a pipe is set to output_level=’document’ :
- Every Sentence Embedding will generate 1 Embedding per Document/row in the input Dataframe, instead of 1 embedding per sentence.
- Every Classifier will classify an entire Document/row
- Each row in the output DF is a 1 to 1 mapping of the original input DF. 1 to 1 mapping from input to output.
- If a pipe is set to output_level=’sentence’ :
- Every Sentence Embedding will generate 1 Embedding per Sentence,
- Every Classifier will classify exactly one sentence
- Each row in the output DF can is mapped to one row in the input DF, but one row in the input DF can have multiple corresponding rows in the output DF. 1 to N mapping from input to output.
- If a pipe is set to output_level=’document’ :
- Improved generation of column names for classifiers. based on input nlu reference
- Improved generation of column names for embeddings, based on input nlu reference
- Improved automatic output level inference
- Various test updates
- Integration of CI pipeline with Github Actions
New Documentation is out!
Check it out here : https://nlp.johnsnowlabs.com/
NLU Version 1.0.1
NLU 1.0.1 Bugfixes
- Fixed bug that caused NER pipelines to crash in NLU when input string caused the NER model to predict without additional metadata
NLU Version 1.0.0
- Automatic to Numpy conversion of embeddings
- Added various testing classes
- New 6 embeddings at once notebook with t-SNE and Medium article
- Integration of Spark NLP 2.6.2 enhancements and bugfixes https://github.com/JohnSnowLabs/spark-nlp/releases/tag/2.6.2
- Updated old T-SNE notebooks with more elegant and simpler generation of t-SNE embeddings
NLU Version 0.2.1
- Various bugfixes
- Improved output column names when using multiple classifirs at once
NLU Version 0.2.0
- Improved output column names classifiers
NLU Version 0.1.0
We are glad to announce that NLU 0.1 has been released! NLU makes the 350+ models and annotators in Spark NLPs arsenal available in just 1 line of python code and it works with Pandas dataframes! A picture says more than a 1000 words, so here is a demo clip of the 12 coolest features in NLU, all just in 1 line!
NLU in action

What does NLU 0.1 include?
- NLU provides everything a data scientist might want to wish for in one line of code!
- 350 + pre-trained models
- 100+ of the latest NLP word embeddings ( BERT, ELMO, ALBERT, XLNET, GLOVE, BIOBERT, ELECTRA, COVIDBERT) and different variations of them
- 50+ of the latest NLP sentence embeddings ( BERT, ELECTRA, USE) and different variations of them
- 50+ Classifiers (NER, POS, Emotion, Sarcasm, Questions, Spam)
- 40+ Supported Languages
- Labeled and Unlabeled Dependency parsing
- Various Text Cleaning and Pre-Processing methods like Stemming, Lemmatizing, Normalizing, Filtering, Cleaning pipelines and more
NLU 0.1 Features Google Collab Notebook Demos
- Named Entity Recognition (NER)
- Part of speech (POS)
- Word and Sentence Embeddings
- BERT Word Embeddings and T-SNE plotting
- BERT Sentence Embeddings and T-SNE plotting
- ALBERT Word Embeddings and T-SNE plotting
- ELMO Word Embeddings and T-SNE plotting
- XLNET Word Embeddings and T-SNE plotting
- ELECTRA Word Embeddings and T-SNE plotting
- COVIDBERT Word Embeddings and T-SNE plotting
- BIOBERT Word Embeddings and T-SNE plotting
- GLOVE Word Embeddings and T-SNE plotting
- USE Sentence Embeddings and T-SNE plotting
- Depenency Parsing
- Text Pre Processing and Cleaning
- Chunkers
- Matchers