PDF processing
Next section describes the transformers that deal with PDF files with the purpose of extracting text and image data from PDF files.
PdfToText
PDFToText extracts text from selectable PDF (with text layout).
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the PDF document |
| originCol | string | path | path to the original file |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| splitPage | bool | true | Whether it needed to split document to pages |
| textStripper | TextStripperType.PDF_TEXT_STRIPPER | Extract unstructured text | |
| sort | bool | false | Sort text during extraction with TextStripperType.PDF_LAYOUT_STRIPPER |
| partitionNum | int | 0 | Force repartition dataframe if set to value more than 0. |
| onlyPageNum | bool | false | Extract only page numbers. |
| extractCoordinates | bool | false | Extract coordinates and store to the positions column |
| storeSplittedPdf | bool | false | Store one page pdf’s for process it using PdfToImage. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | extracted text |
| pageNumCol | string | pagenum | page number or 0 when splitPage = false |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
pdfPath = "path to pdf with text layout"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
transformer = PdfToText() \
.setInputCol("content") \
.setOutputCol("text") \
.setPageNumCol("pagenum") \
.setSplitPage(True)
data = transformer.transform(df)
data.select("pagenum", "text").show()
import com.johnsnowlabs.ocr.transformers.PdfToText
val pdfPath = "path to pdf with text layout"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val transformer = new PdfToText()
.setInputCol("content")
.setOutputCol("text")
.setPageNumCol("pagenum")
.setSplitPage(true)
val data = transformer.transform(df)
data.select("pagenum", "text").show()
Output:
+-------+----------------------+
|pagenum|text |
+-------+----------------------+
|0 |This is a page. |
|1 |This is another page. |
|2 |Yet another page. |
+-------+----------------------+
PdfToImage
PdfToImage renders PDF to an image. To be used with scanned PDF documents.
Output dataframe contains total_pages field with total number of pages.
For process pdf with a big number of pages prefer to split pdf by setting splitNumBatch param.
Number of partitions should be equal to number of cores/executors.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | binary representation of the PDF document |
| originCol | string | path | path to the original file |
| fallBackCol | string | text | extracted text from previous method for detect if need to run transformer as fallBack |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| splitPage | bool | true | whether it needed to split document to pages |
| minSizeBeforeFallback | int | 10 | minimal count of characters to extract to decide, that the document is the PDF with text layout |
| imageType | ImageType | ImageType.TYPE_BYTE_GRAY |
type of the image |
| resolution | int | 300 | Output image resolution in dpi |
| keepInput | boolean | false | Keep input column in dataframe. By default it is dropping. |
| partitionNum | int | 0 | Number of Spark RDD partitions (0 value - without repartition) |
| binarization | boolean | false | Enable/Disable binarization image after extract image. |
| binarizationParams | Array[String] | null | Array of Binarization params in key=value format. |
| splitNumBatch | int | 0 | Number of partitions or size of partitions, related to the splitting strategy. |
| partitionNumAfterSplit | int | 0 | Number of Spark RDD partitions after splitting pdf document (0 value - without repartition). |
| splittingStategy | SplittingStrategy | SplittingStrategy.FIXED_SIZE_OF_PARTITION |
Controls how a single document is split into a number of partitions each containing a number of pages from the original document. This is useful to process documents with high page count. It can be one of {FIXED_SIZE_OF_PARTITION, FIXED_NUMBER_OF_PARTITIONS}, when FIXED_SIZE_OF_PARTITION is used, splitNumBatch represents the size of each partition, and when FIXED_NUMBER_OF_PARTITIONS is used, splitNumBatch represents the number of partitions. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | extracted image struct (Image schema) |
| pageNumCol | string | pagenum | page number or 0 when splitPage = false |
Example:
from sparkocr.transformers import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdfToImage = PdfToImage() \
.setInputCol("content") \
.setOutputCol("text") \
.setPageNumCol("pagenum") \
.setSplitPage(True)
data = pdfToImage.transform(df)
data.select("pagenum", "text").show()
import com.johnsnowlabs.ocr.transformers.PdfToImage
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToImage = new PdfToImage()
.setInputCol("content")
.setOutputCol("text")
.setPageNumCol("pagenum")
.setSplitPage(true)
val data = pdfToImage.transform(df)
data.select("pagenum", "text").show()
ImageToPdf
ImageToPdf transform image to Pdf document.
If dataframe contains few records for same origin path, it groups image by origin
column and create multipage PDF document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | content | binary representation of the PDF document |
Example:
Read images and store them as single page PDF documents.
from sparkocr.transformers import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
# Define transformer for convert to Image struct
binaryToImage = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for store to PDF
imageToPdf = ImageToPdf() \
.setInputCol("image") \
.setOutputCol("content")
# Call transformers
image_df = binaryToImage.transform(df)
pdf_df = pdfToImage.transform(image_df)
pdf_df.select("content").show()
import com.johnsnowlabs.ocr.transformers._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read.format("binaryFile").load(imagePath)
// Define transformer for convert to Image struct
val binaryToImage = new BinaryToImage()
.setInputCol("content")
.setOutputCol("image")
// Define transformer for store to PDF
val imageToPdf = new ImageToPdf()
.setInputCol("image")
.setOutputCol("content")
// Call transformers
val image_df = binaryToImage.transform(df)
val pdf_df = pdfToImage.transform(image_df)
pdf_df.select("content").show()
TextToPdf
TextToPdf renders ocr results to PDF document as text layout. Each symbol will render to the same position
with the same font size as in original image or PDF.
If dataframe contains few records for same origin path, it groups image by origin
column and create multipage PDF document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | positions | column with positions struct |
| inputImage | string | image | image struct (Image schema) |
| inputText | string | text | column name with recognized text |
| originCol | string | path | path to the original file |
| inputContent | string | content | column name with binary representation of original PDF file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | binary representation of the PDF document |
Example:
Read PDF document, run OCR and render results to PDF document.
from sparkocr.transformers import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_image = PdfToImage() \
.setInputCol("content") \
.setOutputCol("image_raw")
binarizer = ImageBinarizer() \
.setInputCol("image_raw") \
.setOutputCol("image") \
.setThreshold(130)
ocr = ImageToText() \
.setInputCol("image") \
.setOutputCol("text") \
.setIgnoreResolution(False) \
.setPageSegMode(PageSegmentationMode.SPARSE_TEXT) \
.setConfidenceThreshold(60)
textToPdf = TextToPdf() \
.setInputCol("positions") \
.setInputImage("image") \
.setOutputCol("pdf")
pipeline = PipelineModel(stages=[
pdf_to_image,
binarizer,
ocr,
textToPdf
])
result = pipeline.transform(df).collect()
# Store to file for debug
with open("test.pdf", "wb") as file:
file.write(result[0].pdf)
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers._
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToImage = new PdfToImage()
.setInputCol("content")
.setOutputCol("image_raw")
.setResolution(400)
val binarizer = new ImageBinarizer()
.setInputCol("image_raw")
.setOutputCol("image")
.setThreshold(130)
val ocr = new ImageToText()
.setInputCol("image")
.setOutputCol("text")
.setIgnoreResolution(false)
.setPageSegMode(PageSegmentationMode.SPARSE_TEXT)
.setConfidenceThreshold(60)
val textToPdf = new TextToPdf()
.setInputCol("positions")
.setInputImage("image")
.setOutputCol("pdf")
val pipeline = new Pipeline()
pipeline.setStages(Array(
pdfToImage,
binarizer,
ocr,
textToPdf
))
val modelPipeline = pipeline.fit(df)
val pdf = modelPipeline.transform(df)
val pdfContent = pdf.select("pdf").collect().head.getAs[Array[Byte]](0)
// store to file
val tmpFile = Files.createTempFile(suffix=".pdf").toAbsolutePath.toString
val fos = new FileOutputStream(tmpFile)
fos.write(pdfContent)
fos.close()
println(tmpFile)
PdfAssembler
PdfAssembler group single page PDF documents by the filename and assemble
muliplepage PDF document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | page_pdf | binary representation of the PDF document |
| originCol | string | path | path to the original file |
| pageNumCol | string | pagenum | for compatibility with another transformers |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | binary representation of the PDF document |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_image = PdfToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setKeepInput(True)
# Run OCR and render results to PDF
ocr = ImageToTextPdf() \
.setInputCol("image") \
.setOutputCol("pdf_page")
# Assemble multipage PDF
pdf_assembler = PdfAssembler() \
.setInputCol("pdf_page") \
.setOutputCol("pdf")
pipeline = PipelineModel(stages=[
pdf_to_image,
ocr,
pdf_assembler
])
pdf = pipeline.transform(df)
pdfContent = pdf.select("pdf").collect().head.getAs[Array[Byte]](0)
# store pdf to file
with open("test.pdf", "wb") as file:
file.write(pdfContent[0].pdf)
import java.io.FileOutputStream
import java.nio.file.Files
import com.johnsnowlabs.ocr.transformers._
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdf_to_image = new PdfToImage()
.setInputCol("content")
.setOutputCol("image")
.setKeepInput(True)
// Run OCR and render results to PDF
val ocr = new ImageToTextPdf()
.setInputCol("image")
.setOutputCol("pdf_page")
// Assemble multipage PDF
val pdf_assembler = new PdfAssembler()
.setInputCol("pdf_page")
.setOutputCol("pdf")
// Create pipeline
val pipeline = new Pipeline()
.setStages(Array(
pdf_to_image,
ocr,
pdf_assembler
))
val pdf = pipeline.fit(df).transform(df)
val pdfContent = pdf.select("pdf").collect().head.getAs[Array[Byte]](0)
// store to pdf file
val tmpFile = Files.createTempFile("with_regions_", s".pdf").toAbsolutePath.toString
val fos = new FileOutputStream(tmpFile)
fos.write(pdfContent)
fos.close()
println(tmpFile)
PdfDrawRegions
PdfDrawRegions transformer for drawing regions to Pdf document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | binary representation of the PDF document |
| originCol | string | path | path to the original file |
| inputRegionsCol | string | region | input column which contain regions |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| lineWidth | integer | 1 | line width for draw regions |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | pdf_regions | binary representation of the PDF document |
Example:
from pyspark.ml import Pipeline
from sparkocr.transformers import *
from sparknlp.annotator import *
from sparknlp.base import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_text = PdfToText() \
.setInputCol("content") \
.setOutputCol("text") \
.setPageNumCol("page") \
.setSplitPage(False)
document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
entity_extractor = TextMatcher() \
.setInputCols("sentence", "token") \
.setEntities("./sparkocr/resources/test-chunks.txt", ReadAs.TEXT) \
.setOutputCol("entity")
position_finder = PositionFinder() \
.setInputCols("entity") \
.setOutputCol("coordinates") \
.setPageMatrixCol("positions") \
.setMatchingWindow(10) \
.setPadding(2)
draw = PdfDrawRegions() \
.setInputRegionsCol("coordinates") \
.setOutputCol("pdf_with_regions") \
.setInputCol("content") \
.setLineWidth(1)
pipeline = Pipeline(stages=[
pdf_to_text,
document_assembler,
sentence_detector,
tokenizer,
entity_extractor,
position_finder,
draw
])
pdfWithRegions = pipeline.fit(df).transform(df)
pdfContent = pdfWithRegions.select("pdf_regions").collect().head.getAs[Array[Byte]](0)
# store to pdf to tmp file
with open("test.pdf", "wb") as file:
file.write(pdfContent[0].pdf_regions)
import java.io.FileOutputStream
import java.nio.file.Files
import com.johnsnowlabs.ocr.transformers._
import com.johnsnowlabs.nlp.{DocumentAssembler, SparkAccessor}
import com.johnsnowlabs.nlp.annotators._
import com.johnsnowlabs.nlp.util.io.ReadAs
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToText = new PdfToText()
.setInputCol("content")
.setOutputCol("text")
.setSplitPage(false)
val documentAssembler = new DocumentAssembler()
.setInputCol("text")
.setOutputCol("document")
val sentenceDetector = new SentenceDetector()
.setInputCols(Array("document"))
.setOutputCol("sentence")
val tokenizer = new Tokenizer()
.setInputCols(Array("sentence"))
.setOutputCol("token")
val entityExtractor = new TextMatcher()
.setInputCols("sentence", "token")
.setEntities("test-chunks.txt", ReadAs.TEXT)
.setOutputCol("entity")
val positionFinder = new PositionFinder()
.setInputCols("entity")
.setOutputCol("coordinates")
.setPageMatrixCol("positions")
.setMatchingWindow(10)
.setPadding(2)
val pdfDrawRegions = new PdfDrawRegions()
.setInputRegionsCol("coordinates")
// Create pipeline
val pipeline = new Pipeline()
.setStages(Array(
pdfToText,
documentAssembler,
sentenceDetector,
tokenizer,
entityExtractor,
positionFinder,
pdfDrawRegions
))
val pdfWithRegions = pipeline.fit(df).transform(df)
val pdfContent = pdfWithRegions.select("pdf_regions").collect().head.getAs[Array[Byte]](0)
// store to pdf to tmp file
val tmpFile = Files.createTempFile("with_regions_", s".pdf").toAbsolutePath.toString
val fos = new FileOutputStream(tmpFile)
fos.write(pdfContent)
fos.close()
println(tmpFile)
Results:

PdfToTextTable
Extract tables from Pdf document page. Input is a column with binary representation of PDF document. As output generate column with tables and tables text chunks coordinates (rows/cols).
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the PDF document |
| originCol | string | path | path to the original file |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| pageIndex | integer | -1 | Page index to extract Tables. |
| guess | bool | false | A logical indicating whether to guess the locations of tables on each page. |
| method | string | decide | Identifying the prefered method of table extraction: basic, spreadsheet. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | TableContainer | tables | Extracted tables |
Example:
from pyspark.ml import Pipeline
from sparkocr.transformers import *
from sparknlp.annotator import *
from sparknlp.base import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_text_table = PdfToTextTable()
pdf_to_text_table.setInputCol("content")
pdf_to_text_table.setOutputCol("table")
pdf_to_text_table.setPageIndex(1)
pdf_to_text_table.setMethod("basic")
table = pdf_to_text_table.transform(df)
# Show first row
table.select(table["table.chunks"].getItem(1)["chunkText"]).show(1, False)
import java.io.FileOutputStream
import java.nio.file.Files
import com.johnsnowlabs.ocr.transformers._
import com.johnsnowlabs.nlp.{DocumentAssembler, SparkAccessor}
import com.johnsnowlabs.nlp.annotators._
import com.johnsnowlabs.nlp.util.io.ReadAs
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToTextTable = new PdfToTextTable()
.setInputCol("content")
.setOutputCol("table")
.pdf_to_text_table.setPageIndex(1)
.pdf_to_text_table.setMethod("basic")
table = pdfToTextTable.transform(df)
// Show first row
table.select(table["table.chunks"].getItem(1)["chunkText"]).show(1, False)
Output:
+------------------------------------------------------------------+
|table.chunks AS chunks#760[1].chunkText |
+------------------------------------------------------------------+
|[Mazda RX4, 21.0, 6, , 160.0, 110, 3.90, 2.620, 16.46, 0, 1, 4, 4]|
+------------------------------------------------------------------+
DOCX processing
Next section describes the transformers that deal with DOCX files with the purpose of extracting text and table data from it.
DocToText
DocToText extracts text from the DOCX document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the DOCX document |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | extracted text |
| pageNumCol | string | pagenum | for compatibility with another transformers |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
docPath = "path to docx with text layout"
# Read DOCX file as binary file
df = spark.read.format("binaryFile").load(docPath)
transformer = DocToText() \
.setInputCol("content") \
.setOutputCol("text")
data = transformer.transform(df)
data.select("pagenum", "text").show()
import com.johnsnowlabs.ocr.transformers.DocToText
val docPath = "path to docx with text layout"
// Read DOCX file as binary file
val df = spark.read.format("binaryFile").load(docPath)
val transformer = new DocToText()
.setInputCol("content")
.setOutputCol("text")
val data = transformer.transform(df)
data.select("pagenum", "text").show()
DocToTextTable
DocToTextTable extracts table data from the DOCX documents.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the PDF document |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | TableContainer | tables | Extracted tables |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
docPath = "path to docx with text layout"
# Read DOCX file as binary file
df = spark.read.format("binaryFile").load(docPath)
transformer = DocToTextTable() \
.setInputCol("content") \
.setOutputCol("tables")
data = transformer.transform(df)
data.select("tables").show()
import com.johnsnowlabs.ocr.transformers.DocToTextTable
val docPath = "path to docx with text layout"
// Read DOCX file as binary file
val df = spark.read.format("binaryFile").load(docPath)
val transformer = new DocToTextTable()
.setInputCol("content")
.setOutputCol("tables")
val data = transformer.transform(df)
data.select("tables").show()
DocToPdf
DocToPdf convert DOCX document to PDF document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the DOCX document |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | binary representation of the PDF document |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
docPath = "path to docx with text layout"
# Read DOCX file as binary file
df = spark.read.format("binaryFile").load(docPath)
transformer = DocToPdf() \
.setInputCol("content") \
.setOutputCol("pdf")
data = transformer.transform(df)
data.select("pdf").show()
import com.johnsnowlabs.ocr.transformers.DocToPdf
val docPath = "path to docx with text layout"
// Read DOCX file as binary file
val df = spark.read.format("binaryFile").load(docPath)
val transformer = new DocToPdf()
.setInputCol("content")
.setOutputCol("pdf")
val data = transformer.transform(df)
data.select("pdf").show()
PptToTextTable
PptToTextTable extracts table data from the PPT and PPTX documents.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the PPT document |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | TableContainer | tables | Extracted tables |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
docPath = "path to docx with text layout"
# Read PPT file as binary file
df = spark.read.format("binaryFile").load(docPath)
transformer = PptToTextTable() \
.setInputCol("content") \
.setOutputCol("tables")
data = transformer.transform(df)
data.select("tables").show()
import com.johnsnowlabs.ocr.transformers.PptToTextTable
val docPath = "path to docx with text layout"
// Read PPT file as binary file
val df = spark.read.format("binaryFile").load(docPath)
val transformer = new PptToTextTable()
.setInputCol("content")
.setOutputCol("tables")
val data = transformer.transform(df)
data.select("tables").show()
PptToPdf
PptToPdf convert PPT and PPTX documents to PDF document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | binary representation of the PPT document |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | binary representation of the PDF document |
NOTE: For setting parameters use setParamName method.
Example
from sparkocr.transformers import *
docPath = "path to PPT with text layout"
# Read DOCX file as binary file
df = spark.read.format("binaryFile").load(docPath)
transformer = PptToPdf() \
.setInputCol("content") \
.setOutputCol("pdf")
data = transformer.transform(df)
data.select("pdf").show()
import com.johnsnowlabs.ocr.transformers.PptToPdf
val docPath = "path to docx with text layout"
// Read PPT file as binary file
val df = spark.read.format("binaryFile").load(docPath)
val transformer = new PptToPdf()
.setInputCol("content")
.setOutputCol("pdf")
val data = transformer.transform(df)
data.select("pdf").show()
Dicom processing
DicomToImage
DicomToImage transforms dicom object (loaded as binary file) to image struct.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | binary dicom object |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | extracted image struct (Image schema) |
| pageNumCol | integer | pagenum | page (image) number begin from 0 |
| metadataCol | string | metadata | Output column name for dicom metatdata ( json formatted ) |
Scala example:
from sparkocr.transformers import *
dicomPath = "path to dicom files"
# Read dicom file as binary file
df = spark.read.format("binaryFile").load(dicomPath)
dicomToImage = DicomToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setMetadataCol("meta")
data = dicomToImage.transform(df)
data.select("image", "pagenum", "meta").show()
import com.johnsnowlabs.ocr.transformers.DicomToImage
val dicomPath = "path to dicom files"
// Read dicom file as binary file
val df = spark.read.format("binaryFile").load(dicomPath)
val dicomToImage = new DicomToImage()
.setInputCol("content")
.setOutputCol("image")
.setMetadataCol("meta")
val data = dicomToImage.transform(df)
data.select("image", "pagenum", "meta").show()
ImageToDicom
ImageToDicom transforms image to Dicom document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| originCol | string | path | path to the original file |
| metadataCol | string | metadata | dicom metatdata ( json formatted ) |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | dicom | binary dicom object |
Scala example:
from sparkocr.transformers import *
imagePath = "path to image file"
# Read image file as binary file
df = spark.read.format("binaryFile").load(imagePath)
binaryToImage = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
image_df = binaryToImage.transform(df)
imageToDicom = ImageToDicom() \
.setInputCol("image") \
.setOutputCol("dicom")
data = imageToDicom.transform(image_df)
data.select("dicom").show()
import com.johnsnowlabs.ocr.transformers.ImageToDicom
val imagePath = "path to image file"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val imageToDicom = new ImageToDicom()
.setInputCol("image")
.setOutputCol("dicom")
val data = imageToDicom.transform(df)
data.select("dicom").show()
DicomToImageV2
Used to convert the dicom frames into images.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Specifies the column containing the DICOM file path or buffer input |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether input columns should be retained in the resulting DataFrame |
| PageNumCol | string | - | Specifies the column containing the page number in the resulting DataFrame |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | Specifies the column containing the images in the resulting DataFrame |
Example:
from sparkocr.transformers import DicomToImageV2
from sparkocr.utils import display_images
df = spark.read.format("binaryFile").load(path_to_dicom_file)
dicom_to_image_v2 = DicomToImageV2() \
.setInputCols(["path"]) \
.setKeepInput(True) \
.setOutputCol("image_raw") \
.setPageNumCol("pagenum")
result = dicom_to_image_v2.transform(df).cache()
display_images(result, "image_raw")
import com.johnsnowlabs.ocr.transformers.DicomToImageV2
import com.johnsnowlabs.ocr.utils.display_images
val df = spark.read.format("binaryFile").load(path_to_dicom_file)
val dicom_to_image_v2 = DicomToImageV2()
.setInputCols(["path"])
.setKeepInput(True)
.setOutputCol("image_raw")
.setPageNumCol("pagenum")
val result = dicom_to_image_v2.transform(df).cache()
val display_images(result, "image_raw")
DicomToImageV3
Used to convert the dicom frames into images. Faster than DicomToImageV2.
Input Columns
| Param name | Type | Default | Description |
|---|---|---|---|
| keepInput | boolean | False | Indicates whether input columns should be retained in the resulting DataFrame |
| pageNumCol | string | - | Specifies the column containing the page number in the resulting DataFrame |
| scale | float | 1 | The scale factor by which the image will be rescaled. Default is ‘1’ (no scaling). |
| frameLimit | int | 0 | Limits the number of frames extracted; set to 0 to extract all frames. |
| compressionThreshold | int | 1 | File size threshold in MB that triggers compression when compressionMode is "auto". |
| compressionMode | string ("auto" | "enabled" | "disabled") |
disabled | Compression behavior: "enabled" compresses every file using compressionQuality; "disabled" never compresses; "auto" compresses only if file size ≥ compressionThreshold. |
| compressionQuality | int (1–95) | 85 | JPEG quality used when compressing (higher = better quality/larger size). |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether input columns should be retained in the resulting DataFrame |
| PageNumCol | string | - | Specifies the column containing the page number in the resulting DataFrame |
| Scale | boolean | 0 | The desired width of the input image, to which the image will be resized |
| FrameLimit | int | 0 | Limits the number of frames extracted; set to 0 to extract all frames. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specifies the column in the output DataFrame that contains the raw images |
Example:
from sparkocr.transformers import DicomSplitter, DicomToImageV3
from sparkocr.enums import SplittingStrategy
from pyspark.ml import PipelineModel
df = spark.read.format("binaryFile").load(dicom_file_path).drop("content")
dicom_splitter = DicomSplitter() \
.setInputCol("path") \
.setOutputCol("frames") \
.setKeepInput(True) \
.setSplitNumBatch(2) \
.setPartitionNum(10) \
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
dicom_to_image = DicomToImageV3() \
.setInputCols(["path", "frames"]) \
.setOutputCol("image_raw") \
.setKeepInput(False) \
.setPageNumCol("page_number") \
.setScale(1) \
.setFrameLimit(0)
pipeline = PipelineModel(stages=[
dicom_splitter,
dicom_to_image
])
result = pipeline.transform(df).cache()
import com.johnsnowlabs.ocr.transformers.DicomSplitter
import com.johnsnowlabs.ocr.transformers.DicomToImageV3
import com.johnsnowlabs.ocr.enums.SplittingStrategy
import com.pyspark.ml.PipelineModel
val df = spark.read.format("binaryFile").load(dicom_file_path).drop("content")
val dicom_splitter = DicomSplitter()
.setInputCol("path")
.setOutputCol("frames")
.setKeepInput(True)
.setSplitNumBatch(2)
.setPartitionNum(10)
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
val dicom_to_image = DicomToImageV3()
.setInputCols(Array("path", "frames"))
.setOutputCol("image_raw")
.setKeepInput(False)
.setPageNumCol("page_number")
.setScale(1)
.setFrameLimit(0)
val pipeline = new PipelineModel().setStages(Array(
dicom_splitter,
dicom_to_image
))
val result = pipeline.transform(df).cache()
DicomDrawRegions
DicomDrawRegions draw regions to Dicom Image.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Binary dicom object |
| inputRegionsCol | string | regions | Detected Array[Coordinates] from PositionFinder |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| scaleFactor | float | 1.0 | Scaling factor for regions. |
| rotated | boolean | False | Enable/Disable support for rotated rectangles |
| keepInput | boolean | False | Keep the original input column |
| compression | string | RLELossless | Compression type |
| forceCompress | boolean | False | True - Force compress image. False - compress only if original image was compressed |
| aggCols | Array[string] | [‘path’] | Sets the columns to be included in aggregation. These columns are preserved in the output DataFrame after transformations |
| forceOutput | boolean | False | Enables decompressed output for DICOM files when compression is unsupported or format constraints prevent re-compression |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | Modified Dicom file data |
Example:
from sparkocr.transformers import *
dicomPath = "path to dicom files"
# Read dicom file as binary file
df = spark.read.format("binaryFile").load(dicomPath)
dicomToImage = DicomToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setMetadataCol("meta")
position_finder = PositionFinder() \
# Usually chunks are created using the deidentification_nlp_pipeline
.setInputCols("ner_chunk") \
.setOutputCol("coordinates") \
.setPageMatrixCol("positions") \
.setPadding(0)
draw_regions = DicomDrawRegions() \
.setInputCol("content") \
.setInputRegionsCol("coordinates") \
.setOutputCol("dicom") \
.setKeepInput(True) \
.setScaleFactor(1/3.0) \
.setAggCols(["path", "content"])
data = dicomToImage.transform(df)
data.select("content", "dicom").show()
// Note: DicomDrawRegions class is not available in the Scala API
// This class is used in the Python API for DICOM image manipulation and transformation.
DicomSplitter
DicomSplitter splits the dicom file into several frames.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Binary dicom object |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| PartitionNum | Int | 5 | The number of partitions in the output DataFrame |
| SplitNumBatch | Int | 2 | The number of frames per row for the input DICOM file |
| KeepInput | boolean | False | Indicates whether the input columns should be included in the result DataFrame |
| SplittingStrategy | enum | FixedSizePartition | Defines the partitioning strategy, either a fixed number of partitions (evenly dividing data across a set number of partitions) or a fixed size of partitions (each partition containing a specified number of elements) |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | The output column name containing the DICOM file’s image frames |
Example:
from sparkocr.transformers import DicomSplitter
from sparkocr.enums import SplittingStrategy
df = spark.read.format("binaryFile").load(dicom_file_path).drop("content")
dicom_splitter = DicomSplitter() \
.setInputCol("path") \
.setOutputCol("frames") \
.setKeepInput(True) \
.setSplitNumBatch(2) \
.setPartitionNum(10) \
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
result = dicom_splitter.transform(df).cache()
import com.johnsnowlabs.ocr.transformers.DicomSplitter
import com.johnsnowlabs.ocr.enums.SplittingStrategy
val df = spark.read.format("binaryFile").load(dicom_file_path).drop("content")
val dicom_splitter = DicomSplitter()
.setInputCol("path")
.setOutputCol("frames")
.setKeepInput(True)
.setSplitNumBatch(2)
.setPartitionNum(10)
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
val result = dicom_splitter.transform(df).cache()
DicomMetadataDeidentifier
Remove Protected Health Information from dicom tags.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCols | array[string] | array[content] | Expects the path or buffer for the DICOM file, along with optional metadata. |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether the input columns should be included in the result DataFrame |
| PlaceHolderText | string | anonymous | Specifies the placeholder text used to deidentify sensitive PHI (Protected Health Information) in DICOM tags |
| RemovePrivateTags | boolean | False | Indicates whether private tags should be removed from the DICOM file |
| BlackList | array[string] | - | Contains a list of specific DICOM tags to be deidentified; only tags in this list will be processed for deidentification if provided |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Column that stores the final DICOM file after de-identifying DICOM tags |
Example:
from sparkocr.transformers import DicomMetadataDeidentifier
df = spark.read.format("binaryFile").load(dicom_file_path)
dicom_deidentifier = DicomMetadataDeidentifier() \
.setInputCols(["path"]) \
.setOutputCol("dicom_deidentified") \
.setKeepInput(True) \
.setPlaceholderText("***") \
.setRemovePrivateTags(True)
result = dicom_deidentifier.transform(df).cache()
result.write.format("binaryFormat") \
.option("type", "dicom") \
.option("field", "dicom_deidentified") \
.option("extension", "dcm") \
.option("prefix", "de-id-ybr") \
.mode("overwrite") \
.save("/content/dicom")
import com.johnsnowlabs.ocr.transformers.DicomMetadataDeidentifier
val df = spark.read.format("binaryFile").load(dicom_file_path)
val dicom_deidentifier = DicomMetadataDeidentifier()
.setInputCols(Array("path"))
.setOutputCol("dicom_deidentified")
.setKeepInput(True)
.setPlaceholderText("***")
.setRemovePrivateTags(True)
val result = dicom_deidentifier.transform(df).cache()
val result.write.format("binaryFormat")
.option("type", "dicom")
.option("field", "dicom_deidentified")
.option("extension", "dcm")
.option("prefix", "de-id-ybr")
.mode("overwrite")
.save("/content/dicom")
DicomDeidentifier
Uses metadata and detected text positions to locate and de-identify sensitive information in both pixel data and metadata.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCols | array[string] | array[content] | Accepts the positions and dicom metadata as input. |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether the input columns should be included in the result DataFrame |
| BlackList | array[string] | anonymous | Specifies a list of words to be removed from pixels. |
| BlackListFile | string | False | Path to CSV file containing a list of words to be removed from pixels. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specifies the column containing the dicom in the resulting DataFrame. |
Example:
from sparkocr.transformers import DicomToMetadata, DicomSplitter, DicomToImageV3, ImageTextDetector, ImageToTextV3, DicomDeidentifier
from sparkocr.enums import OcrOutputFormat, SplittingStrategy
from pyspark.ml import PipelineModel
df = spark.read.format("binaryFile").load(path_to_dicom_file).drop("content")
dicom_to_meta = DicomToMetadata() \
.setInputCol("path") \
.setOutputCol("metadata") \
.setKeepInput(True)
dicom_splitter = DicomSplitter() \
.setInputCol("path") \
.setOutputCol("frames") \
.setKeepInput(True) \
.setSplitNumBatch(2) \
.setPartitionNum(10) \
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
dicom_to_image = DicomToImageV3() \
.setInputCols(["path", "frames"]) \
.setOutputCol("image_raw") \
.setKeepInput(True)
text_detector = ImageTextDetector.pretrained("image_text_detector_opt", "en", "clinical/ocr") \
.setInputCol("image_raw") \
.setOutputCol("text_regions") \
.setSizeThreshold(10) \
.setScoreThreshold(0.9) \
.setLinkThreshold(0.4) \
.setTextThreshold(0.2)
ocr = ImageToTextV3() \
.setInputCols(["image_raw", "text_regions"]) \
.setOutputCol("text")
dicom_deidentifier = DicomDeidentifier() \
.setInputCols(["positions","metadata"]) \
.setKeepInput(True) \
.setOutputCol("entity") \
.setBlackList(["acc"])
pipeline = PipelineModel(stages=[
dicom_to_meta,
dicom_splitter,
dicom_to_image,
text_detector,
ocr,
dicom_deidentifier
])
result = pipeline.transform(df).cache()
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers.DicomToMetadata
import com.johnsnowlabs.ocr.transformers.DicomSplitter
import com.johnsnowlabs.ocr.transformers.DicomToImageV3
import com.johnsnowlabs.ocr.transformers.ImageTextDetector
import com.johnsnowlabs.ocr.transformers.ImageToTextV3
import com.johnsnowlabs.ocr.transformers.DicomDeidentifier
import com.johnsnowlabs.ocr.enums.OcrOutputFormat
import com.johnsnowlabs.ocr.enums.SplittingStrategy
val df = spark.read.format("binaryFile").load(path_to_dicom_file).drop("content")
val dicom_to_meta = DicomToMetadata()
.setInputCol("path")
.setOutputCol("metadata")
.setKeepInput(True)
val dicom_splitter = DicomSplitter()
.setInputCol("path")
.setOutputCol("frames")
.setKeepInput(True)
.setSplitNumBatch(2)
.setPartitionNum(10)
.setSplittingStategy(SplittingStrategy.FIXED_SIZE_OF_PARTITION)
val dicom_to_image = DicomToImageV3()
.setInputCols(Array("path", "frames"))
.setOutputCol("image_raw")
.setKeepInput(True)
val text_detector = ImageTextDetector.pretrained("image_text_detector_opt", "en", "clinical/ocr")
.setInputCol(Array("image_raw"))
.setOutputCol("text_regions")
.setSizeThreshold(10)
.setScoreThreshold(0.9)
.setLinkThreshold(0.4)
.setTextThreshold(0.2)
val ocr = ImageToTextV3()
.setInputCols(Array("image_raw", "text_regions"))
.setOutputCol("text")
val dicom_deidentifier = DicomDeidentifier()
.setInputCols(Array("positions","metadata"))
.setKeepInput(True)
.setOutputCol("entity")
.setBlackList(Array("acc"))
pipeline = new PipelineModel().setStages(Array(
dicom_to_meta,
dicom_splitter,
dicom_to_image,
text_detector,
ocr,
dicom_deidentifier
))
val result = pipeline.transform(df).cache()
DicomToMetadata
Extract metadata from Dicom files.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Specifies the path or buffer for the DICOM file. |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates if the DICOM input columns should remain in the final result DataFrame. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specify the column name to contain the dicom metadata as output |
Example:
from sparkocr.transformers import DicomToMetadata
dicom_to_meta = DicomToMetadata() \
.setInputCol("path") \
.setOutputCol("metadata") \
.setKeepInput(True)
df_dicom = spark.read.format("binaryFile").load(dicom_file_path)
result = dicom_to_meta.transform(df_dicom).cache()
dicom_metadata = result.select("metadata").collect()[0].asDict()["metadata"]
import com.johnsnowlabs.ocr.transformers.DicomToMetadata
val dicom_to_meta = DicomToMetadata()
.setInputCol("path")
.setOutputCol("metadata")
.setKeepInput(True)
val df_dicom = spark.read.format("binaryFile").load(dicom_file_path)
val result = dicom_to_meta.transform(df_dicom).cache()
val dicom_metadata = result.select("metadata").collect()[0].asDict()Array("metadata")
DicomToPDF
Convert Dicom Files to PDF files.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCols | array[string] | - | Specifies the input path or buffer for the DICOM file. |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Determines if the original input should be retained in the resulting DataFrame. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specifies the column name containing the final PDF file resulting from the conversion. |
Example:
from sparkocr.transformers import DicomToPdf, PdfToImage
from pyspark.ml import PipelineModel
df_dicom = spark.read.format("binaryFile").load(dicom_file_path)
dicom_to_pdf = DicomToPdf() \
.setInputCols(["path"]) \
.setOutputCol("dicom_pdf") \
.setKeepInput(True)
pdf_to_image = PdfToImage() \
.setInputCol("dicom_pdf") \
.setOutputCol("image")
pipeline = PipelineModel(stages=[
dicom_to_pdf,
pdf_to_image
])
result = pipeline.transform(df_dicom).cache()
import com.johnsnowlabs.ocr.transformers.DicomToPdf
import com.johnsnowlabs.ocr.transformers.PdfToImage
import com.pyspark.ml.PipelineModel
df_dicom = spark.read.format("binaryFile").load(dicom_file_path)
val dicom_to_pdf = DicomToPdf()
.setInputCols(Array("path"))
.setOutputCol("dicom_pdf")
.setKeepInput(True)
val pdf_to_image = PdfToImage()
.setInputCol("dicom_pdf")
.setOutputCol("image")
val pipeline = new PipelineModel().setStages(Array(
dicom_to_pdf,
pdf_to_image
))
val result = pipeline.transform(df_dicom).cache()
DicomUpdatePDF
Converts pdf file into dicom as a last step for de-identification process.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Specifies the column containing the DICOM file path or buffer input |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether input columns should be retained in the resulting DataFrame |
| InputPdfCol | string | - | Specifies the column containing the pdf object in the resulting DataFrame. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specifies the column containing the dicom in the resulting DataFrame. |
Example:
from sparkocr.transformers import DicomUpdatePdf
dciom_update_pdf = DicomUpdatePdf() \
.setInputCol("path") \
.setInputPdfCol("pdf") \
.setOutputCol("dicom") \
.setKeepInput(True)
result = dciom_update_pdf.transform(df).cache()
result.write.format("binaryFormat") \
.option("type", "dicom") \
.option("field", "dicom") \
.option("extension", "dcm") \
.option("prefix", "de-id-ybr") \
.mode("overwrite") \
.save("/content/dicom")
import com.johnsnowlabs.ocr.transformers.DicomUpdatePdf
val dciom_update_pdf = DicomUpdatePdf()
.setInputCol("path")
.setInputPdfCol("pdf")
.setOutputCol("dicom")
.setKeepInput(True)
val result = dciom_update_pdf.transform(df).cache()
val result.write.format("binaryFormat")
.option("type", "dicom")
.option("field", "dicom")
.option("extension", "dcm")
.option("prefix", "de-id-ybr")
.mode("overwrite")
.save("/content/dicom")
DicomUpdatePDF
Converts pdf file into dicom as a last step for de-identification process.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | Specifies the column containing the DICOM file path or buffer input |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| KeepInput | boolean | False | Indicates whether input columns should be retained in the resulting DataFrame |
| InputPdfCol | string | - | Specifies the column containing the pdf object in the resulting DataFrame. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | - | Specifies the column containing the dicom in the resulting DataFrame. |
Example:
from sparkocr.transformers import DicomUpdatePdf
dciom_update_pdf = DicomUpdatePdf() \
.setInputCol("path") \
.setInputPdfCol("pdf") \
.setOutputCol("dicom") \
.setKeepInput(True)
result = dciom_update_pdf.transform(df).cache()
result.write.format("binaryFormat") \
.option("type", "dicom") \
.option("field", "dicom") \
.option("extension", "dcm") \
.option("prefix", "de-id-ybr") \
.mode("overwrite") \
.save("/content/dicom")
import com.johnsnowlabs.ocr.transformers.DicomUpdatePdf
val dciom_update_pdf = DicomUpdatePdf()
.setInputCol("path")
.setInputPdfCol("pdf")
.setOutputCol("dicom")
.setKeepInput(True)
val result = dciom_update_pdf.transform(df).cache()
val result.write.format("binaryFormat")
.option("type", "dicom")
.option("field", "dicom")
.option("extension", "dcm")
.option("prefix", "de-id-ybr")
.mode("overwrite")
.save("/content/dicom")
Image pre-processing
Next section describes the transformers for image pre-processing: scaling, binarization, skew correction, etc.
BinaryToImage
BinaryToImage transforms image (loaded as binary file) to image struct.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | content | binary representation of the image |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image | extracted image struct (Image schema) |
Scala example:
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read.format("binaryFile").load(imagePath)
binaryToImage = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
data = binaryToImage.transform(df)
data.select("image").show()
import com.johnsnowlabs.ocr.transformers.BinaryToImage
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read.format("binaryFile").load(imagePath)
val binaryToImage = new BinaryToImage()
.setInputCol("content")
.setOutputCol("image")
val data = binaryToImage.transform(df)
data.select("image").show()
GPUImageTransformer
GPUImageTransformer allows to run image pre-processing operations on GPU.
It supports the following operations:
- Scaling
- Otsu thresholding
- Huang thresholding
- Erosion
- Dilation
GPUImageTransformer allows to add few operations. To add operations you need to call
one of the methods with params:
| Method name | Params | Description |
|---|---|---|
| addScalingTransform | factor | Scale image by scaling factor. |
| addOtsuTransform | The automatic thresholder utilizes the Otsu threshold method. | |
| addHuangTransform | The automatic thresholder utilizes the Huang threshold method. | |
| addDilateTransform | width, height | Computes the local maximum of a pixels rectangular neighborhood. The rectangles size is specified by its half-width and half-height. |
| addErodeTransform | width, height | Computes the local minimum of a pixels rectangular neighborhood. The rectangles size is specified by its half-width and half-height |
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| imageType | ImageType | ImageType.TYPE_BYTE_BINARY |
Type of the output image |
| gpuName | string | ”” | GPU device name. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | transformed_image | image struct (Image schema) |
Example:
from sparkocr.transformers import *
from sparkocr.enums import ImageType
from sparkocr.utils import display_images
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
transformer = GPUImageTransformer() \
.setInputCol("image") \
.setOutputCol("transformed_image") \
.addHuangTransform() \
.addScalingTransform(3) \
.addDilateTransform(2, 2) \
.setImageType(ImageType.TYPE_BYTE_BINARY)
pipeline = PipelineModel(stages=[
binary_to_image,
transformer
])
result = pipeline.transform(df)
display_images(result, "transformed_image")
import com.johnsnowlabs.ocr.transformers.GPUImageTransformer
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new GPUImageTransformer()
.setInputCol("image")
.setOutputCol("transformed_image")
.addHuangTransform()
.addScalingTransform(3)
.addDilateTransform(2, 2)
.setImageType(ImageType.TYPE_BYTE_BINARY)
val data = transformer.transform(df)
data.storeImage("transformed_image")
ImageBinarizer
ImageBinarizer transforms image to binary color schema, based on threshold.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| threshold | int | 170 |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | binarized_image | image struct (Image schema) |
Example:
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath) \
.asImage("image")
binirizer = ImageBinarizer() \
.setInputCol("image") \
.setOutputCol("binary_image") \
.setThreshold(100)
data = binirizer.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageBinarizer
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val binirizer = new ImageBinarizer()
.setInputCol("image")
.setOutputCol("binary_image")
.setThreshold(100)
val data = binirizer.transform(df)
data.storeImage("binary_image")
Original image:

Binarized image with 100 threshold:

ImageAdaptiveBinarizer
Supported Methods:
- OTSU. Returns a single intensity threshold that separate pixels into two classes, foreground and background.
- Gaussian local thresholding. Thresholds the image using a locally adaptive threshold that is computed using a local square region centered on each pixel. The threshold is equal to the gaussian weighted sum of the surrounding pixels times the scale.
- Sauvola. Is a Local thresholding technique that are useful for images where the background is not uniform.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| width | float | 90 | Width of square region. |
| method | TresholdingMethod | TresholdingMethod.GAUSSIAN |
Method used to determine adaptive threshold. |
| scale | float | 1.1f | Scale factor used to adjust threshold. |
| imageType | ImageType | ImageType.TYPE_BYTE_BINARY |
Type of the output image |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | binarized_image | image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
from sparkocr.utils import display_image
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
adaptive_thresholding = ImageAdaptiveBinarizer() \
.setInputCol("image") \
.setOutputCol("binarized_image") \
.setWidth(100) \
.setScale(1.1)
pipeline = PipelineModel(stages=[
binary_to_image,
adaptive_thresholding
])
result = pipeline.transform(df)
for r in result.select("image", "corrected_image").collect():
display_image(r.image)
display_image(r.corrected_image)
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val binirizer = new ImageAdaptiveBinarizer()
.setInputCol("image")
.setOutputCol("binary_image")
.setWidth(100)
.setScale(1.1)
val data = binirizer.transform(df)
data.storeImage("binary_image")
ImageAdaptiveThresholding
Compute a threshold mask image based on local pixel neighborhood and apply it to image.
Also known as adaptive or dynamic thresholding. The threshold value is the weighted mean for the local neighborhood of a pixel subtracted by a constant.
Supported methods:
- GAUSSIAN
- MEAN
- MEDIAN
- WOLF
- SINGH
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| blockSize | int | 170 | Odd size of pixel neighborhood which is used to calculate the threshold value (e.g. 3, 5, 7, …, 21, …). |
| method | AdaptiveThresholdingMethod | AdaptiveThresholdingMethod.GAUSSIAN | Method used to determine adaptive threshold for local neighbourhood in weighted mean image. |
| offset | int | Constant subtracted from weighted mean of neighborhood to calculate the local threshold value. Default offset is 0. | |
| mode | string | The mode parameter determines how the array borders are handled, where cval is the value when mode is equal to ‘constant’ | |
| cval | int | Value to fill past edges of input if mode is ‘constant’. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | binarized_image | image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
from sparkocr.utils import display_image
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
adaptive_thresholding = ImageAdaptiveThresholding() \
.setInputCol("scaled_image") \
.setOutputCol("binarized_image") \
.setBlockSize(21) \
.setOffset(73)
pipeline = PipelineModel(stages=[
binary_to_image,
adaptive_thresholding
])
result = pipeline.transform(df)
for r in result.select("image", "corrected_image").collect():
display_image(r.image)
display_image(r.corrected_image)
// Implemented only for Python
Original image:

Binarized image:

ImageScaler
ImageScaler scales image by provided scale factor or needed output size.
It supports keeping original ratio of image by padding the image in case fixed output size.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| scaleFactor | double | 1.0 | scale factor |
| keepRatio | boolean | false | Keep original ratio of image |
| width | int | 0 | Output width of image |
| height | int | 0 | Outpu height of imgae |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | scaled_image | scaled image struct (Image schema) |
Example:
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath) \
.asImage("image")
transformer = ImageScaler() \
.setInputCol("image") \
.setOutputCol("scaled_image") \
.setScaleFactor(0.5)
data = transformer.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageScaler
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageScaler()
.setInputCol("image")
.setOutputCol("scaled_image")
.setScaleFactor(0.5)
val data = transformer.transform(df)
data.storeImage("scaled_image")
ImageAdaptiveScaler
ImageAdaptiveScaler detects font size and scales image for have desired font size.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| desiredSize | int | 34 | desired size of font in pixels |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | scaled_image | scaled image struct (Image schema) |
Example:
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath) \
.asImage("image")
transformer = ImageAdaptiveScaler() \
.setInputCol("image") \
.setOutputCol("scaled_image") \
.setDesiredSize(34)
data = transformer.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageAdaptiveScaler
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageAdaptiveScaler()
.setInputCol("image")
.setOutputCol("scaled_image")
.setDesiredSize(34)
val data = transformer.transform(df)
data.storeImage("scaled_image")
ImageSkewCorrector
ImageSkewCorrector detects skew of the image and rotates it.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| rotationAngle | double | 0.0 | rotation angle |
| automaticSkewCorrection | boolean | true | enables/disables adaptive skew correction |
| halfAngle | double | 5.0 | half the angle(in degrees) that will be considered for correction |
| resolution | double | 1.0 | The step size(in degrees) that will be used for generating correction angle candidates |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | corrected_image | corrected image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
from sparkocr.utils import display_images
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
skew_corrector = ImageSkewCorrector() \
.setInputCol("image") \
.setOutputCol("corrected_image") \
.setAutomaticSkewCorrection(True)
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
skew_corrector
])
data = pipeline.transform(df)
display_images(data, "corrected_image")
import com.johnsnowlabs.ocr.transformers.ImageSkewCorrector
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageSkewCorrector()
.setInputCol("image")
.setOutputCol("corrected_image")
.setAutomaticSkewCorrection(true)
val data = transformer.transform(df)
data.storeImage("corrected_image")
Original image:

Corrected image:

ImageNoiseScorer
ImageNoiseScorer computes noise score for each region.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| inputRegionsCol | string | regions | regions |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| method | NoiseMethod string | NoiseMethod.RATIO | method of computation noise score |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | noisescores | noise score for each region |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
from sparkocr.enums import NoiseMethod
imagePath = "path to image"
# Read image file as binary file
df = spark.read \
.format("binaryFile") \
.load(imagePath) \
.asImage("image")
# Define transformer for detect regions
layoutAnalyzer = ImageLayoutAnalyzer() \
.setInputCol("image") \
.setOutputCol("regions")
# Define transformer for compute noise level for each region
noisescorer = ImageNoiseScorer() \
.setInputCol("image") \
.setOutputCol("noiselevel") \
.setInputRegionsCol("regions") \
.setMethod(NoiseMethod.VARIANCE)
# Define pipeline
pipeline = Pipeline()
pipeline.setStages(Array(
layoutAnalyzer,
noisescorer
))
data = pipeline.transform(df)
data.select("path", "noiselevel").show()
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers.{ImageNoiseScorer, ImageLayoutAnalyzer}
import com.johnsnowlabs.ocr.NoiseMethod
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect regions
val layoutAnalyzer = new ImageLayoutAnalyzer()
.setInputCol("image")
.setOutputCol("regions")
// Define transformer for compute noise level for each region
val noisescorer = new ImageNoiseScorer()
.setInputCol("image")
.setOutputCol("noiselevel")
.setInputRegionsCol("regions")
.setMethod(NoiseMethod.VARIANCE)
// Define pipeline
val pipeline = new Pipeline()
pipeline.setStages(Array(
layoutAnalyzer,
noisescorer
))
val modelPipeline = pipeline.fit(spark.emptyDataFrame)
val data = modelPipeline.transform(df)
data.select("path", "noiselevel").show()
Output:
+------------------+-----------------------------------------------------------------------------+
|path |noiselevel |
+------------------+-----------------------------------------------------------------------------+
|file:./noisy.png |[32.01805641767766, 32.312916551193354, 29.99257352247787, 30.62470388308217]|
+------------------+-----------------------------------------------------------------------------+
ImageRemoveObjects
python only
ImageRemoveObjects to remove background objects.
It supports removing:
- objects less than elements of font with minSizeFont size
- objects less than minSizeObject
- holes less than minSizeHole
- objects more than maxSizeObject
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | None | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| minSizeFont | int | 10 | Min size font in pt. |
| minSizeObject | int | None | Min size of object which will keep on image [*]. |
| connectivityObject | int | 0 | The connectivity defining the neighborhood of a pixel. |
| minSizeHole | int | None | Min size of hole which will keep on image[ *]. |
| connectivityHole | int | 0 | The connectivity defining the neighborhood of a pixel. |
| maxSizeObject | int | None | Max size of object which will keep on image [*]. |
| connectivityMaxObject | int | 0 | The connectivity defining the neighborhood of a pixel. |
[*] : None value disables removing objects.
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | None | scaled image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
remove_objects = ImageRemoveObjects() \
.setInputCol("image") \
.setOutputCol("corrected_image") \
.setMinSizeObject(20)
pipeline = PipelineModel(stages=[
binary_to_image,
remove_objects
])
data = pipeline.transform(df)
// Implemented only for Python
ImageMorphologyOperation
python only
ImageMorphologyOperationis a transformer for applying morphological operations to image.
It supports following operation:
- Erosion
- Dilation
- Opening
- Closing
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | None | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| operation | MorphologyOperationType | MorphologyOperationType.OPENING | Operation type |
| kernelShape | KernelShape | KernelShape.DISK | Kernel shape. |
| kernelSize | int | 1 | Kernel size in pixels. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | None | scaled image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setOperation(MorphologyOperationType.OPENING)
adaptive_thresholding = ImageAdaptiveThresholding() \
.setInputCol("image") \
.setOutputCol("corrected_image") \
.setBlockSize(75) \
.setOffset(0)
opening = ImageMorphologyOperation() \
.setInputCol("corrected_image") \
.setOutputCol("opening_image") \
.setkernelSize(1)
pipeline = PipelineModel(stages=[
binary_to_image,
adaptive_thresholding,
opening
])
result = pipeline.transform(df)
for r in result.select("image", "corrected_image").collect():
display_image(r.image)
display_image(r.corrected_image)
// Implemented only for Python
Original image:

Opening image:

ImageCropper
ImageCropperis a transformer for cropping image.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| cropRectangle | Rectangle | Rectangle(0,0,0,0) | Image rectangle. |
| cropSquareType | CropSquareType | CropSquareType.TOP_LEFT | Type of square. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | cropped_image | scaled image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setOperation(MorphologyOperationType.OPENING)
cropper = ImageCropper() \
.setInputCol("image") \
.setOutputCol("cropped_image") \
.setCropRectangle((0, 0, 200, 110))
pipeline = PipelineModel(stages=[
binary_to_image,
cropper
])
result = pipeline.transform(df)
for r in result.select("image", "cropped_image").collect():
display_image(r.image)
display_image(r.cropped_image)
import com.johnsnowlabs.ocr.transformers.ImageAdaptiveScaler
import com.johnsnowlabs.ocr.OcrContext.implicits._
import java.awt.Rectangle
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val rectangle: Rectangle = new Rectangle(0, 0, 200, 110)
val cropper: ImageCropper = new ImageCropper()
.setInputCol("image")
.setOutputCol("cropped_image")
.setCropRectangle(rectangle)
val data = transformer.transform(df)
data.storeImage("cropped_image")
Splitting image to regions
ImageLayoutAnalyzer
ImageLayoutAnalyzer analyzes the image and determines regions of text.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| pageSegMode | PageSegmentationMode | AUTO | page segmentation mode |
| pageIteratorLevel | PageIteratorLevel | BLOCK | page iteration level |
| ocrEngineMode | EngineMode | LSTM_ONLY | OCR engine mode |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | region | array of [Coordinaties]ocr_structures#coordinate-schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for detect regions
layout_analyzer = ImageLayoutAnalyzer() \
.setInputCol("image") \
.setOutputCol("regions")
pipeline = PipelineModel(stages=[
binary_to_image,
layout_analyzer
])
data = pipeline.transform(df)
data.show()
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers.{ImageSplitRegions, ImageLayoutAnalyzer}
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect regions
val layoutAnalyzer = new ImageLayoutAnalyzer()
.setInputCol("image")
.setOutputCol("regions")
val data = layoutAnalyzer.transform(df)
data.show()
ImageSplitRegions
ImageSplitRegions splits image into regions.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| inputRegionsCol | string | region | array of [Coordinaties]ocr_structures#coordinate-schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| explodeCols | Array[string] | Columns which need to explode | |
| rotated | boolean | False | Support rotated regions |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | region_image | image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for detect regions
layout_analyzer = ImageLayoutAnalyzer() \
.setInputCol("image") \
.setOutputCol("regions")
splitter = ImageSplitRegions()
.setInputCol("image")
.setRegionCol("regions")
.setOutputCol("region_image")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
layout_analyzer,
splitter
])
data = pipeline.transform(df)
data.show()
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers.{ImageSplitRegions, ImageLayoutAnalyzer}
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect regions
val layoutAnalyzer = new ImageLayoutAnalyzer()
.setInputCol("image")
.setOutputCol("regions")
val splitter = new ImageSplitRegions()
.setInputCol("image")
.setRegionCol("regions")
.setOutputCol("region_image")
// Define pipeline
val pipeline = new Pipeline()
pipeline.setStages(Array(
layoutAnalyzer,
splitter
))
val modelPipeline = pipeline.fit(spark.emptyDataFrame)
val data = pipeline.transform(df)
data.show()
ImageDrawAnnotations
ImageDrawAnnotations draw annotations with label and score to the image.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| inputChunksCol | string | region | array of Annotation |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| lineWidth | Int | 4 | Line width for draw rectangles |
| fontSize | Int | 12 | Font size for render labels and score |
| rectColor | Color | Color.black | Color of lines |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image_with_chunks | image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToHocr() \
.setInputCol("image") \
.setOutputCol("hocr")
tokenizer = HocrTokenizer() \
.setInputCol("hocr") \
.setOutputCol("token")
draw_annotations = ImageDrawAnnotations() \
.setInputCol("image") \
.setInputChunksCol("token") \
.setOutputCol("image_with_annotations") \
.setFilledRect(False) \
.setFontSize(40) \
.setRectColor(Color.red)
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr,
tokenizer,
image_with_annotations
])
result = pipeline.transform(df)
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val imageToHocr = new ImageToHocr()
.setInputCol("image")
.setOutputCol("hocr")
val tokenizer = new HocrTokenizer()
.setInputCol("hocr")
.setOutputCol("token")
val draw_annotations = new ImageDrawAnnotations()
.setInputCol("image")
.setInputChunksCol("token")
.setOutputCol("image_with_annotations")
.setFilledRect(False)
.setFontSize(40)
.setRectColor(Color.red)
val pipeline = new Pipeline()
pipeline.setStages(Array(
imageToHocr,
tokenizer,
draw_annotations
))
val modelPipeline = pipeline.fit(df)
val result = modelPipeline.transform(df)
ImageDrawRegions
ImageDrawRegions draw regions with label and score to the image.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| inputRegionsCol | string | region | array of [Coordinaties]ocr_structures#coordinate-schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| lineWidth | Int | 4 | Line width for draw rectangles |
| fontSize | Int | 12 | Font size for render labels and score |
| rotated | boolean | False | Support rotated regions |
| rectColor | Color | Color.black | Color outline for bounding box |
| filledRect | boolean | False | Enable/Disable filling rectangle |
| sourceImageHeightCol | Int | height_dimension | Original annotation reference height |
| sourceImageWidthCol | Int | width_dimension | Original annotation reference width |
| scaleBoundingBoxes | Boolean | True | sourceImage height & width are required for scaling. Necessary to ensure accurate regions despite image transformations. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | image_with_regions | image struct (Image schema) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
from sparkocr.enums import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read.format("binaryFile").load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for detect regions
layout_analyzer = ImageLayoutAnalyzer() \
.setInputCol("image") \
.setOutputCol("regions")
draw = ImageDrawRegions() \
.setInputCol("image") \
.setRegionCol("regions") \
.setRectColor(Color.red) \
.setOutputCol("image_with_regions")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
layout_analyzer,
draw
])
data = pipeline.transform(df)
data.show()
import org.apache.spark.ml.Pipeline
import java.awt.Color
import com.johnsnowlabs.ocr.transformers.{ImageSplitRegions, ImageLayoutAnalyzer}
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read.format("binaryFile").load(imagePath).asImage("image")
// Define transformer for detect regions
val layoutAnalyzer = new ImageLayoutAnalyzer()
.setInputCol("image")
.setOutputCol("regions")
val draw = new ImageDrawRegions()
.setInputCol("image")
.setRegionCol("regions")
.setRectColor(Color.RED)
.setOutputCol("image_with_regions")
// Define pipeline
val pipeline = new Pipeline()
pipeline.setStages(Array(
layoutAnalyzer,
draw
))
val modelPipeline = pipeline.fit(spark.emptyDataFrame)
val data = pipeline.transform(df)
data.show()
Characters recognition
Next section describes the estimators for OCR
ImageToText
ImageToText runs OCR for input image, return recognized text
to outputCol and positions with font size to ‘positionsCol’ column.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| pageSegMode | PageSegmentationMode | AUTO | page segmentation mode |
| pageIteratorLevel | PageIteratorLevel | BLOCK | page iteration level |
| ocrEngineMode | EngineMode | LSTM_ONLY | OCR engine mode |
| language | Language | Language.ENG | language |
| confidenceThreshold | int | 0 | Confidence threshold. |
| ignoreResolution | bool | false | Ignore resolution from metadata of image. |
| ocrParams | array of strings | [] | Array of Ocr params in key=value format. |
| pdfCoordinates | bool | false | Transform coordinates in positions to PDF points. |
| modelData | string | Path to the local model data. | |
| modelType | ModelType | ModelType.BASE | Model type |
| downloadModelData | bool | false | Download model data from JSL S3 |
| withSpaces | bool | false | Include spaces to output positions. |
| keepLayout | bool | false | Keep layout of text at result. |
| outputSpaceCharacterWidth | int | 8 | Output space character width in pts for layout keeper. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | Recognized text |
| positionsCol | string | positions | Positions of each block of text (related to pageIteratorLevel) in PageMatrix |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToText() \
.setInputCol("image") \
.setOutputCol("text") \
.setOcrParams(["preserve_interword_spaces=1", ])
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr
])
data = pipeline.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageToText
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageToText()
.setInputCol("image")
.setOutputCol("text")
.setOcrParams(Array("preserve_interword_spaces=1"))
val data = transformer.transform(df)
print(data.select("text").collect()[0].text)
Image:

Output:
FOREWORD
Electronic design engineers are the true idea men of the electronic
industries. They create ideas and use them in their designs, they stimu-
late ideas in other designers, and they borrow and adapt ideas from
others. One could almost say they feed on and grow on ideas.
ImageToTextV2
ImageToTextV2 is based on the transformers architecture, and combines CV and NLP
in one model. It is a visual encoder-decoder model. The Encoder is based on ViT,
and the decoder on RoBERTa model.
ImageToTextV2 can work on CPU, but GPU is preferred in order to achieve acceptable performance.
ImageToTextV2 can receive regions representing single line texts, or regions coming from a text detection model.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCols | Array[string] | [Pipeline components] | Can use as input image struct (Image schema) and regions. |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| lineTolerance | integer | 15 | Line tolerance in pixels. It’s used for grouping text regions by lines. |
| borderWidth | integer | 5 | A value of more than 0 enables to border text regions with width equal to the value of the parameter. |
| spaceWidth | integer | 10 | A value of more than 0 enables to add white spaces between words on the image. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | text | Recognized text |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
text_detector = ImageTextDetectorV2 \
.pretrained("image_text_detector_v2", "en", "clinical/ocr") \
.setInputCol("image") \
.setOutputCol("text_regions") \
.setWithRefiner(True) \
.setSizeThreshold(20)
ocr = ImageToTextV2.pretrained("ocr_base_printed", "en", "clinical/ocr") \
.setInputCols(["image", "text_regions"]) \
.setOutputCol("text")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
text_detector,
ocr
])
data = pipeline.transform(df)
data.show()
not implemented
Image:
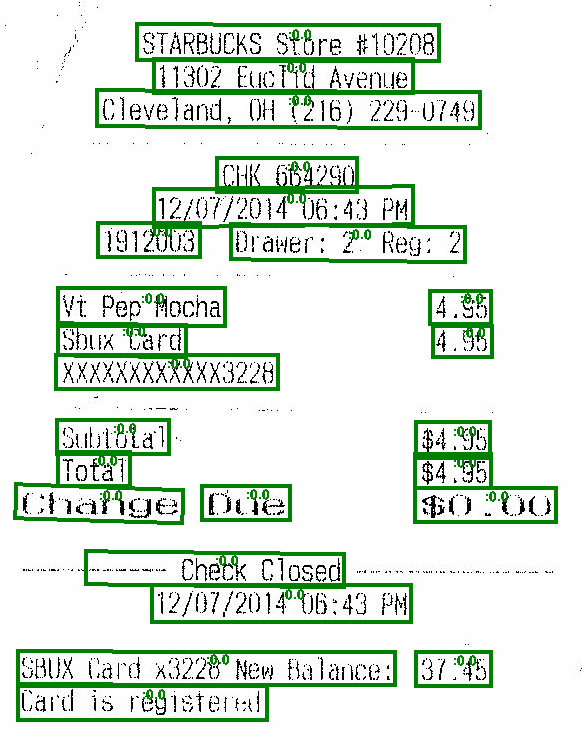
Output:
STARBUCKS STORE #10208
11302 EUCLID AVENUE
CLEVELAND, OH (216) 229-0749
CHK 664290
12/07/2014 06:43 PM
1912003 DRAWER: 2. REG: 2
VT PEP MOCHA 4.95
SBUX CARD 4.95
XXXXXXXXXXXX3228
SUBTOTAL $4.95
TOTAL $4.95
CHANGE DUE $0.00
---- CHECK CLOSED
12/07/2014 06:43 PM
SBUX CARD X3228 NEW BALANCE: 37.45
CARD IS REGISTERED
ImageToTextPdf
ImageToTextPdf runs OCR for input image, render recognized text to
the PDF as an invisible text layout with an original image.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| originCol | string | path | path to the original file |
| pageNumCol | string | pagenum | for compatibility with another transformers |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| ocrParams | array of strings | [] | Array of Ocr params in key=value format. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | Recognized text rendered to PDF |
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToTextPdf() \
.setInputCol("image") \
.setOutputCol("pdf")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr
])
data = pipeline.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageToTextPdf()
.setInputCol("image")
.setOutputCol("pdf")
val data = transformer.transform(df)
data.show()
ImageToHocr
ImageToHocr runs OCR for input image, return recognized text and bounding boxes
to outputCol column in HOCR format.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| pageSegMode | PageSegmentationMode | AUTO | page segmentation mode |
| pageIteratorLevel | PageIteratorLevel | BLOCK | page iteration level |
| ocrEngineMode | EngineMode | LSTM_ONLY | OCR engine mode |
| language | string | eng | language |
| ignoreResolution | bool | true | Ignore resolution from metadata of image. |
| ocrParams | array of strings | [] | Array of Ocr params in key=value format. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | hocr | Recognized text |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToHocr() \
.setInputCol("image") \
.setOutputCol("hocr")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr
])
data = pipeline.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageToHocr
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageToHocr()
.setInputCol("image")
.setOutputCol("hocr")
val data = transformer.transform(df)
print(data.select("hocr").collect()[0].hocr)
Image:

Output:
<div class='ocr_page' id='page_1' title='image ""; bbox 0 0 1280 467; ppageno 0'>
<div class='ocr_carea' id='block_1_1' title="bbox 516 80 780 114">
<p class='ocr_par' id='par_1_1' lang='eng' title="bbox 516 80 780 114">
<span class='ocr_line' id='line_1_1' title="bbox 516 80 780 114; baseline 0 -1; x_size 44; x_descenders 11; x_ascenders 11">
<span class='ocrx_word' id='word_1_1' title='bbox 516 80 780 114; x_wconf 96'>FOREWORD</span>
</span>
</p>
</div>
<div class='ocr_carea' id='block_1_2' title="bbox 40 237 1249 425">
<p class='ocr_par' id='par_1_2' lang='eng' title="bbox 40 237 1249 425">
<span class='ocr_line' id='line_1_2' title="bbox 122 237 1249 282; baseline 0.001 -12; x_size 45; x_descenders 12; x_ascenders 13">
<span class='ocrx_word' id='word_1_2' title='bbox 122 237 296 270; x_wconf 96'>Electronic</span>
<span class='ocrx_word' id='word_1_3' title='bbox 308 237 416 281; x_wconf 96'>design</span>
<span class='ocrx_word' id='word_1_4' title='bbox 428 243 588 282; x_wconf 96'>engineers</span>
<span class='ocrx_word' id='word_1_5' title='bbox 600 250 653 271; x_wconf 96'>are</span>
<span class='ocrx_word' id='word_1_6' title='bbox 665 238 718 271; x_wconf 96'>the</span>
<span class='ocrx_word' id='word_1_7' title='bbox 731 246 798 272; x_wconf 97'>true</span>
<span class='ocrx_word' id='word_1_8' title='bbox 810 238 880 271; x_wconf 96'>idea</span>
<span class='ocrx_word' id='word_1_9' title='bbox 892 251 963 271; x_wconf 96'>men</span>
<span class='ocrx_word' id='word_1_10' title='bbox 977 238 1010 272; x_wconf 96'>of</span>
<span class='ocrx_word' id='word_1_11' title='bbox 1021 238 1074 271; x_wconf 96'>the</span>
<span class='ocrx_word' id='word_1_12' title='bbox 1086 239 1249 272; x_wconf 96'>electronic</span>
</span>
<span class='ocr_line' id='line_1_3' title="bbox 41 284 1248 330; baseline 0.002 -13; x_size 44; x_descenders 11; x_ascenders 12">
<span class='ocrx_word' id='word_1_13' title='bbox 41 284 214 318; x_wconf 96'>industries.</span>
<span class='ocrx_word' id='word_1_14' title='bbox 227 284 313 328; x_wconf 96'>They</span>
<span class='ocrx_word' id='word_1_15' title='bbox 324 292 427 319; x_wconf 96'>create</span>
<span class='ocrx_word' id='word_1_16' title='bbox 440 285 525 319; x_wconf 96'>ideas</span>
<span class='ocrx_word' id='word_1_17' title='bbox 537 286 599 318; x_wconf 96'>and</span>
<span class='ocrx_word' id='word_1_18' title='bbox 611 298 668 319; x_wconf 96'>use</span>
<span class='ocrx_word' id='word_1_19' title='bbox 680 286 764 319; x_wconf 96'>them</span>
<span class='ocrx_word' id='word_1_20' title='bbox 777 291 808 319; x_wconf 96'>in</span>
<span class='ocrx_word' id='word_1_21' title='bbox 821 286 900 319; x_wconf 96'>their</span>
<span class='ocrx_word' id='word_1_22' title='bbox 912 286 1044 330; x_wconf 96'>designs,</span>
<span class='ocrx_word' id='word_1_23' title='bbox 1058 286 1132 330; x_wconf 93'>they</span>
<span class='ocrx_word' id='word_1_24' title='bbox 1144 291 1248 320; x_wconf 92'>stimu-</span>
</span>
<span class='ocr_line' id='line_1_4' title="bbox 42 332 1247 378; baseline 0.002 -14; x_size 44; x_descenders 12; x_ascenders 12">
<span class='ocrx_word' id='word_1_25' title='bbox 42 332 103 364; x_wconf 97'>late</span>
<span class='ocrx_word' id='word_1_26' title='bbox 120 332 204 365; x_wconf 96'>ideas</span>
<span class='ocrx_word' id='word_1_27' title='bbox 223 337 252 365; x_wconf 96'>in</span>
<span class='ocrx_word' id='word_1_28' title='bbox 271 333 359 365; x_wconf 96'>other</span>
<span class='ocrx_word' id='word_1_29' title='bbox 376 333 542 377; x_wconf 96'>designers,</span>
<span class='ocrx_word' id='word_1_30' title='bbox 561 334 625 366; x_wconf 96'>and</span>
<span class='ocrx_word' id='word_1_31' title='bbox 643 334 716 377; x_wconf 96'>they</span>
<span class='ocrx_word' id='word_1_32' title='bbox 734 334 855 366; x_wconf 96'>borrow</span>
<span class='ocrx_word' id='word_1_33' title='bbox 873 334 934 366; x_wconf 96'>and</span>
<span class='ocrx_word' id='word_1_34' title='bbox 954 335 1048 378; x_wconf 96'>adapt</span>
<span class='ocrx_word' id='word_1_35' title='bbox 1067 334 1151 367; x_wconf 96'>ideas</span>
<span class='ocrx_word' id='word_1_36' title='bbox 1169 334 1247 367; x_wconf 96'>from</span>
</span>
<span class='ocr_line' id='line_1_5' title="bbox 40 379 1107 425; baseline 0.002 -13; x_size 45; x_descenders 12; x_ascenders 12">
<span class='ocrx_word' id='word_1_37' title='bbox 40 380 151 412; x_wconf 96'>others.</span>
<span class='ocrx_word' id='word_1_38' title='bbox 168 383 238 412; x_wconf 96'>One</span>
<span class='ocrx_word' id='word_1_39' title='bbox 252 379 345 412; x_wconf 96'>could</span>
<span class='ocrx_word' id='word_1_40' title='bbox 359 380 469 413; x_wconf 96'>almost</span>
<span class='ocrx_word' id='word_1_41' title='bbox 483 392 537 423; x_wconf 96'>say</span>
<span class='ocrx_word' id='word_1_42' title='bbox 552 381 626 424; x_wconf 96'>they</span>
<span class='ocrx_word' id='word_1_43' title='bbox 641 381 712 414; x_wconf 96'>feed</span>
<span class='ocrx_word' id='word_1_44' title='bbox 727 393 767 414; x_wconf 96'>on</span>
<span class='ocrx_word' id='word_1_45' title='bbox 783 381 845 414; x_wconf 96'>and</span>
<span class='ocrx_word' id='word_1_46' title='bbox 860 392 945 425; x_wconf 97'>grow</span>
<span class='ocrx_word' id='word_1_47' title='bbox 959 393 999 414; x_wconf 96'>on</span>
<span class='ocrx_word' id='word_1_48' title='bbox 1014 381 1107 414; x_wconf 95'>ideas.</span>
</span>
</p>
</div>
</div>
ImageBrandsToText
ImageBrandsToText runs OCR for specified brands of input image, return recognized text
to outputCol and positions with font size to ‘positionsCol’ column.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| pageSegMode | PageSegmentationMode | AUTO | page segmentation mode |
| pageIteratorLevel | PageIteratorLevel | BLOCK | page iteration level |
| ocrEngineMode | EngineMode | LSTM_ONLY | OCR engine mode |
| language | string | eng | language |
| confidenceThreshold | int | 0 | Confidence threshold. |
| ignoreResolution | bool | true | Ignore resolution from metadata of image. |
| ocrParams | array of strings | [] | Array of Ocr params in key=value format. |
| brandsCoords | string | Json with coordinates of brands. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | structure | image_brands | Structure with recognized text from brands. |
| textCol | string | text | Recognized text |
| positionsCol | string | positions | Positions of each block of text (related to pageIteratorLevel) |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageBrandsToText() \
.setInputCol("image") \
.setOutputCol("text") \
.setBrandsCoords("""[
{
"name":"part_one",
"rectangle":{
"x":286,
"y":65,
"width":542,
"height":342
}
},
{
"name":"part_two",
"rectangle":{
"x":828,
"y":65,
"width":1126,
"height":329
}
}
]""")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr
])
data = pipeline.transform(df)
data.show()
import com.johnsnowlabs.ocr.transformers.ImageToText
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val transformer = new ImageBrandsToText()
.setInputCol("image")
.setOutputCol("text")
.setBrandsCoordsStr(
"""
[
{
"name":"part_one",
"rectangle":{
"x":286,
"y":65,
"width":542,
"height":342
}
},
{
"name":"part_two",
"rectangle":{
"x":828,
"y":65,
"width":1126,
"height":329
}
}
]
""".stripMargin)
val data = transformer.transform(df)
print(data.select("text").collect()[0].text)
Other
Next section describes the extra transformers
PositionFinder
PositionFinder find the position of input text entities in the original document. This annotator will return coordinates for entities that were extracted from a text column in the same row. It will receive entities in the form of chunks, and positions for characters or words.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCols | string | image | Input annotations columns |
| pageMatrixCol | string | Column name for Page Matrix schema |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| matchingWindow | int | 10 | Textual range to match in context, applies in both direction |
| windowPageTolerance | boolean | true | whether or not to increase tolerance as page number grows |
| padding | int | 5 | padding for area |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | Name of output column for store coordinates. |
Example:
from pyspark.ml import Pipeline
from sparkocr.transformers import *
from sparknlp.annotator import *
from sparknlp.base import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_text = PdfToText() \
.setInputCol("content") \
.setOutputCol("text") \
.setPageNumCol("page") \
.setSplitPage(False)
document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
entity_extractor = TextMatcher() \
.setInputCols("sentence", "token") \
.setEntities("./sparkocr/resources/test-chunks.txt", ReadAs.TEXT) \
.setOutputCol("entity")
position_finder = PositionFinder() \
.setInputCols("entity") \
.setOutputCol("coordinates") \
.setPageMatrixCol("positions") \
.setMatchingWindow(10) \
.setPadding(2)
pipeline = Pipeline(stages=[
pdf_to_text,
document_assembler,
sentence_detector,
tokenizer,
entity_extractor,
position_finder
])
results = pipeline.fit(df).transform(df)
results.show()
import com.johnsnowlabs.ocr.transformers._
import com.johnsnowlabs.nlp.{DocumentAssembler, SparkAccessor}
import com.johnsnowlabs.nlp.annotators._
import com.johnsnowlabs.nlp.util.io.ReadAs
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToText = new PdfToText()
.setInputCol("content")
.setOutputCol("text")
.setSplitPage(false)
val documentAssembler = new DocumentAssembler()
.setInputCol("text")
.setOutputCol("document")
val sentenceDetector = new SentenceDetector()
.setInputCols(Array("document"))
.setOutputCol("sentence")
val tokenizer = new Tokenizer()
.setInputCols(Array("sentence"))
.setOutputCol("token")
val entityExtractor = new TextMatcher()
.setInputCols("sentence", "token")
.setEntities("test-chunks.txt", ReadAs.TEXT)
.setOutputCol("entity")
val positionFinder = new PositionFinder()
.setInputCols("entity")
.setOutputCol("coordinates")
.setPageMatrixCol("positions")
.setMatchingWindow(10)
.setPadding(2)
// Create pipeline
val pipeline = new Pipeline()
.setStages(Array(
pdfToText,
documentAssembler,
sentenceDetector,
tokenizer,
entityExtractor,
positionFinder
))
val results = pipeline.fit(df).transform(df)
results.show()
Algorithm
The algorithm for PositionFinder is pretty simple; while processing the list of entities to be matched, the sequence of positions is consumed by the algorithm as a stream, to try to determine the coordinates for each entity.
A few details:
- The chunks and the char/word positions must be in the same order, as the algorithm alternatively “consumes” elements from both positions and chunks.
- The algorithm does backtracking, meaning that if some chunk entity was not matched when the end of the positions stream was reached, the process continues with the next entity exactly where the last entity was matched in the positions stream.
- As stated befire, positions can be char-level or word-level, according to the stages that were used to create the input to PositionFinder.
- Normalization of the source text(over which entities are extracted) can make the PositionFinder fail, as the entities will be normalized as well, but the ‘positions’, i.e., the individual coordinates for each character don’t reflect this normalization.
- Matching is not strictly textual, you can think of the matching piece of the algorithm like an automaton that will receive inputs(pieces of words, words, or characters) and will “accept” at some point and return a coordinate(or more if, for example, the entity is multi-line). Some soft matching is applied like relaxing the match for punctuation, spaces, or newlines.
- Multi-line entities, spawning two lines will return more than one coordinate in general, you can use output coordinate metadata, and chunk_id, to match coordinates to entities in this situation.
- When for some reason there are entities that couldn’t be matched you will see an error log like this,
ERROR PositionFinder: PositionFinder unmatched:::Annotation(type: chunk, begin: 47, end: 70, result: Cristiano Ronaldo)
In that example, a name entity couldn’t be matched.
UpdateTextPosition
UpdateTextPosition update output text and keep old coordinates of original document.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | positions | Сolumn name with original positions struct |
| InputText | string | replace_text | Column name for New Text to replace Old one |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | output_positions | Name of output column for updated positions struct. |
Example:
from pyspark.ml import Pipeline
from sparkocr.transformers import *
from sparknlp.annotator import *
from sparknlp.base import *
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_text = PdfToText() \
.setInputCol("content") \
.setOutputCol("text") \
.setPageNumCol("page") \
.setSplitPage(False)
document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("tokens")
spell = NorvigSweetingModel().pretrained("spellcheck_norvig", "en") \
.setInputCols("tokens") \
.setOutputCol("spell")
tokenAssem = TokenAssembler() \
.setInputCols("spell") \
.setOutputCol("newDocs")
updatedText = UpdateTextPosition() \
.setInputCol("positions") \
.setOutputCol("output_positions") \
.setInputText("newDocs.result")
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
spell,
tokenAssem,
updatedText
])
results = pipeline.fit(df).transform(df)
results.show()
import com.johnsnowlabs.nlp.annotators.Tokenizer
import com.johnsnowlabs.nlp.annotators.sbd.pragmatic.SentenceDetector
import com.johnsnowlabs.nlp.annotators.spell.norvig.NorvigSweetingModel
import com.johnsnowlabs.nlp.{DocumentAssembler, TokenAssembler}
import com.johnsnowlabs.ocr.transformers._
import org.apache.spark.ml.Pipeline
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToText = new PdfToText()
.setInputCol("content")
.setOutputCol("text")
val documentAssembler = new DocumentAssembler()
.setInputCol("text")
.setOutputCol("document")
val sentence = new SentenceDetector()
.setInputCols("document")
.setOutputCol("sentence")
val token = new Tokenizer()
.setInputCols("document")
.setOutputCol("tokens")
val spell = NorvigSweetingModel.pretrained("spellcheck_norvig", "en")
.setInputCols("tokens")
.setOutputCol("spell")
val tokenAssem = new TokenAssembler()
.setInputCols("spell")
.setOutputCol("newDocs")
val updatedText = new UpdateTextPosition()
.setInputCol("positions")
.setOutputCol("output_positions")
.setInputText("newDocs.result")
val pipeline = new Pipeline()
.setStages(Array(
pdfToText,
documentAssembler,
sentence,
token,
spell,
tokenAssem,
updatedText
))
val results = pipeline.fit(df).transform(df)
results.show()
FoundationOneReportParser
FoundationOneReportParser is a transformer for parsing FoundationOne reports.
Current implementation supports parsing patient info, genomic, biomarker findings and gene lists
from appendix.
Output format is json.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | text | Сolumn name with text of report |
| originCol | string | path | path to the original file |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | report | Name of output column with report in json format. |
Example:
from pyspark.ml import Pipeline
from sparkocr.transformers import *
from sparkocr.enums import TextStripperType
pdfPath = "path to pdf"
# Read PDF file as binary file
df = spark.read.format("binaryFile").load(pdfPath)
pdf_to_text = PdfToText()
pdf_to_text.setInputCol("content")
pdf_to_text.setOutputCol("text")
pdf_to_text.setSplitPage(False)
pdf_to_text.setTextStripper(TextStripperType.PDF_LAYOUT_TEXT_STRIPPER)
genomic_parser = FoundationOneReportParser()
genomic_parser.setInputCol("text")
genomic_parser.setOutputCol("report")
report = genomic_parser.transform(pdf_to_text.transform(df)).collect()
import com.johnsnowlabs.ocr.transformers._
import org.apache.spark.ml.Pipeline
val pdfPath = "path to pdf"
// Read PDF file as binary file
val df = spark.read.format("binaryFile").load(pdfPath)
val pdfToText = new PdfToText()
.setInputCol("content")
.setOutputCol("text")
.setSplitPage(false)
.setTextStripper(TextStripperType.PDF_LAYOUT_TEXT_STRIPPER)
val genomicsParser = new FoundationOneReportParser()
.setInputCol("text")
.setOutputCol("report")
val pipeline = new Pipeline()
pipeline.setStages(Array(
pdfToText,
genomicsParser
))
val modelPipeline = pipeline.fit(df)
val report = modelPipeline.transform(df)
Output:
{
"Patient" : {
"disease" : "Unknown primary melanoma",
"name" : "Lekavich Gloria",
"date_of_birth" : "11 November 1926",
"sex" : "Female",
"medical_record" : "11111"
},
"Physician" : {
"ordering_physician" : "Genes Pinley",
"medical_facility" : "Health Network Cancer Institute",
"additional_recipient" : "Nath",
"medical_facility_id" : "202051",
"pathologist" : "Manqju Nwath"
},
"Specimen" : {
"specimen_site" : "Rectum",
"specimen_id" : "AVS 1A",
"specimen_type" : "Slide",
"date_of_collection" : "20 March 2015",
"specimen_received" : "30 March 2015 "
},
"Biomarker_findings" : [ {
"name" : "Tumor Mutation Burden",
"state" : "TMB-Low (3Muts/Mb)",
"actionability" : "No therapies or clinical trials. "
} ],
"Genomic_findings" : [ {
"name" : "FLT3",
"state" : "amplification",
"therapies_with_clinical_benefit_in_patient_tumor_type" : [ "none" ],
"therapies_with_clinical_benefit_in_other_tumor_type" : [ "Sorafenib", "Sunitinib", "Ponatinib" ]
}
],
"Appendix" : {
"dna_gene_list" : [ "ABL1", "ACVR1B", "AKT1", .... ],
"dna_gene_list_rearrangement" : [ "ALK", "BCL2", "BCR", .... ],
"additional_assays" : [ "Tumor Mutation Burden (TMB)", "Microsatellite Status (MS)" ]
}
}
HocrDocumentAssembler
HocrDocumentAssembler prepares data into a format that is processable by Spark NLP.
Output Annotator Type: DOCUMENT
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | hocr | Сolumn name with HOCR of the document |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | document | Name of output column. |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToHocr() \
.setInputCol("image") \
.setOutputCol("hocr")
hocr_document_assembler = HocrDocumentAssembler() \
.setInputCol("hocr") \
.setOutputCol("document")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr,
hocr_document_assembler
])
result = pipeline.transform(df)
result.select("document").show()
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val imageToHocr = new ImageToHocr()
.setInputCol("image")
.setOutputCol("hocr")
val hocrDocumentAssembler = HocrDocumentAssembler()
.setInputCol("hocr")
.setOutputCol("document")
val pipeline = new Pipeline()
pipeline.setStages(Array(
imageToHocr,
hocrDocumentAssembler
))
val modelPipeline = pipeline.fit(df)
val result = modelPipeline.transform(df)
result.select("document").show()
Output:
+--------------------------------------------------------------------+
| document |
+--------------------------------------------------------------------+
| [[document, 0, 4392, Patient Nam Financial Numbe Random Hospital...|
+--------------------------------------------------------------------+
HocrTokenizer
HocrTokenizer prepares into a format that is processable by Spark NLP.\
HocrTokenizer puts to metadata coordinates and ocr confidence.
Output Annotator Type: TOKEN
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | hocr | Сolumn name with HOCR of the document. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | token | Name of output column. |
Example:
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
ocr = ImageToHocr() \
.setInputCol("image") \
.setOutputCol("hocr")
tokenizer = HocrTokenizer() \
.setInputCol("hocr") \
.setOutputCol("token")
# Define pipeline
pipeline = PipelineModel(stages=[
binary_to_image,
ocr,
tokenizer
])
result = pipeline.transform(df)
result.select("token").show()
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
val imageToHocr = new ImageToHocr()
.setInputCol("image")
.setOutputCol("hocr")
val tokenizer = HocrTokenizer()
.setInputCol("hocr")
.setOutputCol("token")
val pipeline = new Pipeline()
pipeline.setStages(Array(
imageToHocr,
tokenizer
))
val modelPipeline = pipeline.fit(df)
val result = modelPipeline.transform(df)
result.select("token").show()
Output:
+--------------------------------------------------------------------+
| token |
+--------------------------------------------------------------------+
| [[token, 0, 6, patient, [x -> 2905, y -> 527, height -> 56, |
| confidence -> 95, word -> Patient, width -> 230], []], [token, 8, |
|10, nam, [x -> 3166, y -> 526, height -> 55, confidence -> 95, word |
|-> Nam, width -> 158], []] ... |
+--------------------------------------------------------------------+