ImageTableDetector
ImageTableDetector is a DL model for detecting tables on the image.
It’s based on CascadeTabNet which used Cascade mask Region-based CNN High-Resolution Network (Cascade mask R-CNN HRNet).
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| scoreThreshold | float | 0.9 | Score threshold for output regions. |
| applyCorrection | boolean | false | Enable correction of results. |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | table_regions | array of [Coordinaties]ocr_structures#coordinate-schema) |
Example:
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect tables
val table_detector = ImageTableDetector
.pretrained("general_model_table_detection_v2")
.setInputCol("image")
.setOutputCol("table_regions")
val draw_regions = new ImageDrawRegions()
.setInputCol("image")
.setInputRegionsCol("table_regions")
.setOutputCol("image_with_regions")
pipeline = PipelineModel(stages=[
binary_to_image,
table_detector,
draw_regions
])
val data = pipeline.transform(df)
data.storeImage("image_with_regions")
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for detect tables
table_detector = ImageTableDetector \
.pretrained("general_model_table_detection_v2", "en", "clinical/ocr") \
.setInputCol("image") \
.setOutputCol("table_regions")
draw_regions = ImageDrawRegions() \
.setInputCol("image") \
.setInputRegionsCol("table_regions") \
.setOutputCol("image_with_regions")
pipeline = PipelineModel(stages=[
binary_to_image,
table_detector,
draw_regions
])
data = pipeline.transform(df)
display_images(data, "image_with_regions")
Output:
ImageTableCellDetector
ImageTableCellDetector detect cells in a table image. It’s based on an
image processing algorithm that detects horizontal and vertical lines.
Current implementation support few algorithm for extract cells:
- CellDetectionAlgos.CONTOURS works only for bordered tables.
- CellDetectionAlgos.MORPHOPS works for bordered, borderless and combined tables.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| algoType | CellDetectionAlgos | CellDetectionAlgos.CONTOURS | Algorithm for detect cells. |
| algoParams | string | row_treshold=0.05,row_treshold_wide=1.0, row_min_wide=5,column_treshold=0.05, column_treshold_wide=5,column_min_wide=5 | Parameters of ‘MORPHOPS’ cells detection algorithm |
| drawDetectedLines | boolean | false | Enable to draw detected lines to the output image |
| keepOriginalLines | boolean | false | Keep original images on the output image |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | cells | array of coordinates of cells |
| outputImageCol | string | output_image | output image |
Example:
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect cells
val transformer = new ImageTableCellDetector()
.setInputCol("image")
.setOutputCol("cells")
val data = transformer.transform(df)
data.select("cells").show()
from pyspark.ml import PipelineModel
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image")
# Define transformer for detect cells
transformer = ImageTableCellDetector \
.setInputCol("image") \
.setOutputCol("cells") \
.setAlgoParams("row_treshold=0.05")
pipeline = PipelineModel(stages=[
binary_to_image,
transformer
])
data = pipeline.transform(df)
data.select("cells").show()
Image:
Output:*
+----------------------------------------------------------+
| cells |
+----------------------------------------------------------+
||[[[[15, 17, 224, 53]], [[241, 17, 179, 53]], [[423, 17, |
| 194, 53]], [[619, 17, 164, 53]] .... |
+----------------------------------------------------------+
ImageCellsToTextTable
ImageCellsToTextTable runs OCR for cells regions on image, return recognized text
to outputCol as TableContainer structure.
Input Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| inputCol | string | image | image struct (Image schema) |
| cellsCol | string | celss | Array of cells |
Parameters
| Param name | Type | Default | Description |
|---|---|---|---|
| strip | bool | true | Strip output text. |
| margin | bool | 1 | Margin of cells in pixelx. |
| pageSegMode | PageSegmentationMode | AUTO | page segmentation mode |
| ocrEngineMode | EngineMode | LSTM_ONLY | OCR engine mode |
| language | Language | Language.ENG | language |
| ocrParams | array of strings | [] | Array of Ocr params in key=value format. |
| pdfCoordinates | bool | false | Transform coordinates in positions to PDF points. |
| modelData | string | Path to the local model data. | |
| modelType | ModelType | ModelType.BASE | Model type |
| downloadModelData | bool | false | Download model data from JSL S3 |
| outputFormat | TableOutputFormat | TableOutputFormat.TABLE | Output format |
Output Columns
| Param name | Type | Default | Column Data Description |
|---|---|---|---|
| outputCol | string | table | Recognized text as TableContainer |
Example:
import org.apache.spark.ml.Pipeline
import com.johnsnowlabs.ocr.transformers.*
import com.johnsnowlabs.ocr.OcrContext.implicits._
val imagePath = "path to image"
// Read image file as binary file
val df = spark.read
.format("binaryFile")
.load(imagePath)
.asImage("image")
// Define transformer for detect cells
val cell_detector = new ImageTableCellDetector()
.setInputCol("image")
.setOutputCol("cells")
val table_recognition = new ImageCellsToTextTable()
.setInputCol("image")
.setOutputCol("tables")
.setMargin(2)
// Define pipeline
val pipeline = new Pipeline()
pipeline.setStages(Array(
cell_detector,
table_recognition
))
val modelPipeline = pipeline.fit(spark.emptyDataFrame)
val results = modelPipeline.transform(df)
results.select("tables")
.withColumn("cells", explode(col("tables.chunks")))
.select((0 until 7).map(i => col("cells")(i).getField("chunkText").alias(s"col$i")): _*)
.show(false)
from pyspark.ml import PipelineModel
import pyspark.sql.functions as f
from sparkocr.transformers import *
imagePath = "path to image"
# Read image file as binary file
df = spark.read
.format("binaryFile")
.load(imagePath)
binary_to_image = BinaryToImage()
binary_to_image.setImageType(ImageType.TYPE_BYTE_GRAY)
binary_to_image.setInputCol("content")
cell_detector = TableCellDetector()
cell_detector.setInputCol("image")
cell_detector.setOutputCol("cells")
cell_detector.setKeepInput(True)
table_recognition = ImageCellsToTextTable()
table_recognition.setInputCol("image")
table_recognition.setCellsCol('cells')
table_recognition.setMargin(2)
table_recognition.setStrip(True)
table_recognition.setOutputCol('table')
pipeline = PipelineModel(stages=[
binary_to_image,
cell_detector,
table_recognition
])
result = pipeline.transform(df)
results.select("table") \
.withColumn("cells", f.explode(f.col("table.chunks"))) \
.select([f.col("cells")[i].getField("chunkText").alias(f"col{i}") for i in
range(0, 7)]) \
.show(20, False)
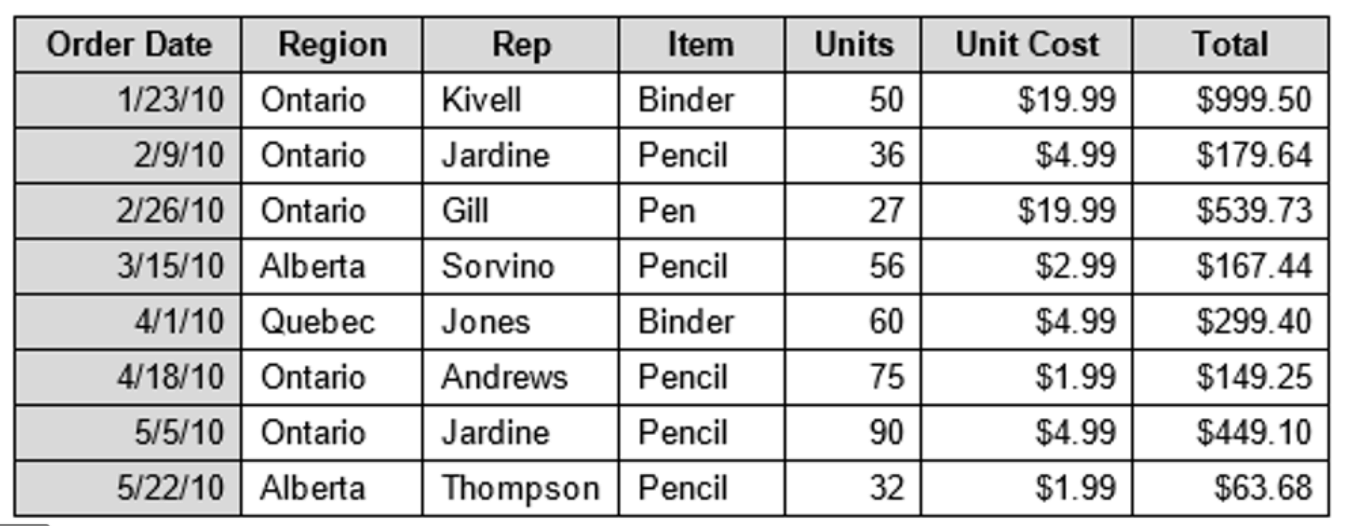
Image:

Output:
+----------+-------+--------+------+-----+---------+-------+
|col0 |col1 |col2 |col3 |col4 |col5 |col6 |
+----------+-------+--------+------+-----+---------+-------+
|Order Date|Region |Rep |Item |Units|Unit Cost|Total |
|1/23/10 |Ontario|Kivell |Binder|50 |$19.99 |$999.50|
|2/9/10 |Ontario|Jardine |Pencil|36 |$4.99 |$179.64|
|2/26/10 |Ontario|Gill |Pen |27 |$19.99 |$539.73|
|3/15/10 |Alberta|Sorvino |Pencil|56 |$2.99 |$167.44|
|4/1/10 |Quebec |Jones |Binder|60 |$4.99 |$299.40|
|4/18/10 |Ontario|Andrews |Pencil|75 |$1.99 |$149.25|
|5/5/10 |Ontario|Jardine |Pencil|90 |$4.99 |$449.10|
|5/22/10 |Alberta|Thompson|Pencil|32 |$1.99 |$63.68 |
+----------+-------+--------+------+-----+---------+-------+
PREVIOUSPipeline components