Annotations can be exported in various format for storage and later use. You can export the annotations applied to the tasks of any project by going to the Export button on the top-right corner of this page. You will be navigated to the Export page and from there you can select the format and configure the export options to export the annotations to a file/s.
Supported Formats for Text Projects
The completions and predictions are stored in a database for fast search and access. Completions and predictions can be exported into the formats described below.
JSON
You can export the manual annotations (completions) and automatic annotations (predictions) to JSON format using the JSON option on the Export page. An example of JSON export file is shown below:
[
{
"completions": [
{
"created_username": "eric",
"created_ago": "2022-10-29T14:42:50.867Z",
"lead_time": 82,
"result": [
{
"value": {
"start": 175,
"end": 187,
"text": "tuberculosis",
"labels": [
"MedicalCondition"
],
"confidence": 0.9524
},
"id": "zgam2AbdmY",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 213,
"end": 239,
"text": "Mycobacterium tuberculosis",
"labels": [
"Pathogen"
],
"confidence": 0.904775
},
"id": "1v76SqlWtj",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 385,
"end": 394,
"text": "pneumonia",
"labels": [
"MedicalCondition"
],
"confidence": 0.91655
},
"id": "CURKae4Eca",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 436,
"end": 449,
"text": "Streptococcus",
"labels": [
"Pathogen"
],
"confidence": 0.9157500000000001
},
"id": "cM5BvAsZL4",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 454,
"end": 465,
"text": "Pseudomonas",
"labels": [
"Pathogen"
],
"confidence": 0.91495
},
"id": "KGOLhb8OPV",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 532,
"end": 540,
"text": "Shigella",
"labels": [
"Pathogen"
],
"confidence": 0.91655
},
"id": "JCIhVQTDZl",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 542,
"end": 555,
"text": "Campylobacter",
"labels": [
"Pathogen"
],
"confidence": 0.9163
},
"id": "CkxrbwvFzb",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 561,
"end": 571,
"text": "Salmonella",
"labels": [
"Pathogen"
],
"confidence": 0.9164000000000001
},
"id": "c6ev6McH4Z",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 623,
"end": 630,
"text": "tetanus",
"labels": [
"MedicalCondition"
],
"confidence": 0.97
},
"id": "9ZmEaJnqKG",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 632,
"end": 645,
"text": "typhoid fever",
"labels": [
"MedicalCondition"
],
"confidence": 0.976675
},
"id": "Uo5CWzdd1S",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 647,
"end": 657,
"text": "diphtheria",
"labels": [
"MedicalCondition"
],
"confidence": 0.9737
},
"id": "7nc71jXT3P",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 659,
"end": 667,
"text": "syphilis",
"labels": [
"MedicalCondition"
],
"confidence": 0.97355
},
"id": "nIKfsOWNyE",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 673,
"end": 689,
"text": "Hansen's disease",
"labels": [
"MedicalCondition"
],
"confidence": 0.899025
},
"id": "SyuVYMn7ax",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 30,
"end": 38,
"text": "bacteria",
"labels": [
"Pathogen"
],
"confidence": 1
},
"id": "lq7qtJj1yX",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 98,
"end": 106,
"text": "bacteria",
"labels": [
"Pathogen"
],
"confidence": 1
},
"id": "kxaB_gMstN",
"from_name": "label",
"to_name": "text",
"type": "labels"
}
],
"honeypot": true,
"copied_from": "prediction: 11001",
"id": 11001,
"confidence_range": [
0,
1
],
"copy": true,
"cid": "11001",
"data_type": "prediction",
"updated_at": "2022-10-29T15:13:03.445569Z",
"updated_by": "eric",
"submitted_at": "2022-10-30T20:57:54.303"
},
{
"created_username": "jenny",
"created_ago": "2022-10-29T15:03:51.669Z",
"lead_time": 0,
"result": [
{
"value": {
"start": 175,
"end": 187,
"text": "tuberculosis",
"labels": [
"MedicalCondition"
],
"confidence": 0.9524
},
"id": "zgam2AbdmY",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 213,
"end": 239,
"text": "Mycobacterium tuberculosis",
"labels": [
"Pathogen"
],
"confidence": 0.904775
},
"id": "1v76SqlWtj",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 385,
"end": 394,
"text": "pneumonia",
"labels": [
"MedicalCondition"
],
"confidence": 0.91655
},
"id": "CURKae4Eca",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 436,
"end": 449,
"text": "Streptococcus",
"labels": [
"Pathogen"
],
"confidence": 0.9157500000000001
},
"id": "cM5BvAsZL4",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 454,
"end": 465,
"text": "Pseudomonas",
"labels": [
"Pathogen"
],
"confidence": 0.91495
},
"id": "KGOLhb8OPV",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 532,
"end": 540,
"text": "Shigella",
"labels": [
"Pathogen"
],
"confidence": 0.91655
},
"id": "JCIhVQTDZl",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 542,
"end": 555,
"text": "Campylobacter",
"labels": [
"Pathogen"
],
"confidence": 0.9163
},
"id": "CkxrbwvFzb",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 561,
"end": 571,
"text": "Salmonella",
"labels": [
"Pathogen"
],
"confidence": 0.9164000000000001
},
"id": "c6ev6McH4Z",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 623,
"end": 630,
"text": "tetanus",
"labels": [
"MedicalCondition"
],
"confidence": 0.97
},
"id": "9ZmEaJnqKG",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 632,
"end": 645,
"text": "typhoid fever",
"labels": [
"MedicalCondition"
],
"confidence": 0.976675
},
"id": "Uo5CWzdd1S",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 647,
"end": 657,
"text": "diphtheria",
"labels": [
"MedicalCondition"
],
"confidence": 0.9737
},
"id": "7nc71jXT3P",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 659,
"end": 667,
"text": "syphilis",
"labels": [
"MedicalCondition"
],
"confidence": 0.97355
},
"id": "nIKfsOWNyE",
"from_name": "label",
"to_name": "text",
"type": "labels"
},
{
"value": {
"start": 673,
"end": 689,
"text": "Hansen's disease",
"labels": [
"MedicalCondition"
],
"confidence": 0.899025
},
"id": "SyuVYMn7ax",
"from_name": "label",
"to_name": "text",
"type": "labels"
}
],
"honeypot": true,
"confidence_range": [
0,
1
],
"submitted_at": "2022-10-29T20:48:51.669",
"id": 11002
}
],
"predictions": [
{
"created_username": "SparkNLP Pre-annotation",
"result": [
{
"from_name": "label",
"id": "zgam2AbdmY",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 187,
"labels": [
"MedicalCondition"
],
"start": 175,
"text": "tuberculosis",
"confidence": "0.9524"
}
},
{
"from_name": "label",
"id": "1v76SqlWtj",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 239,
"labels": [
"Pathogen"
],
"start": 213,
"text": "Mycobacterium tuberculosis",
"confidence": "0.904775"
}
},
{
"from_name": "label",
"id": "CURKae4Eca",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 394,
"labels": [
"MedicalCondition"
],
"start": 385,
"text": "pneumonia",
"confidence": "0.91655"
}
},
{
"from_name": "label",
"id": "cM5BvAsZL4",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 449,
"labels": [
"Pathogen"
],
"start": 436,
"text": "Streptococcus",
"confidence": "0.9157500000000001"
}
},
{
"from_name": "label",
"id": "KGOLhb8OPV",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 465,
"labels": [
"Pathogen"
],
"start": 454,
"text": "Pseudomonas",
"confidence": "0.91495"
}
},
{
"from_name": "label",
"id": "JCIhVQTDZl",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 540,
"labels": [
"Pathogen"
],
"start": 532,
"text": "Shigella",
"confidence": "0.91655"
}
},
{
"from_name": "label",
"id": "CkxrbwvFzb",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 555,
"labels": [
"Pathogen"
],
"start": 542,
"text": "Campylobacter",
"confidence": "0.9163"
}
},
{
"from_name": "label",
"id": "c6ev6McH4Z",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 571,
"labels": [
"Pathogen"
],
"start": 561,
"text": "Salmonella",
"confidence": "0.9164000000000001"
}
},
{
"from_name": "label",
"id": "9ZmEaJnqKG",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 630,
"labels": [
"MedicalCondition"
],
"start": 623,
"text": "tetanus",
"confidence": "0.97"
}
},
{
"from_name": "label",
"id": "Uo5CWzdd1S",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 645,
"labels": [
"MedicalCondition"
],
"start": 632,
"text": "typhoid fever",
"confidence": "0.976675"
}
},
{
"from_name": "label",
"id": "7nc71jXT3P",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 657,
"labels": [
"MedicalCondition"
],
"start": 647,
"text": "diphtheria",
"confidence": "0.9737"
}
},
{
"from_name": "label",
"id": "nIKfsOWNyE",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 667,
"labels": [
"MedicalCondition"
],
"start": 659,
"text": "syphilis",
"confidence": "0.97355"
}
},
{
"from_name": "label",
"id": "SyuVYMn7ax",
"source": "$text",
"to_name": "text",
"type": "labels",
"value": {
"end": 689,
"labels": [
"MedicalCondition"
],
"start": 673,
"text": "Hansen's disease",
"confidence": "0.899025"
}
}
],
"created_ago": "2022-10-29T14:07:58.553246Z",
"id": 11001
}
],
"created_at": "2022-10-29 14:07:12",
"created_by": "admin",
"data": {
"text": "Although the vast majority of bacteria are harmless or beneficial to one's body, a few pathogenic bacteria can cause infectious diseases. The most common bacterial disease is tuberculosis, caused by the bacterium Mycobacterium tuberculosis, which affects about 2 million people mostly in sub-Saharan Africa. Pathogenic bacteria contribute to other globally important diseases, such as pneumonia, which can be caused by bacteria such as Streptococcus and Pseudomonas, and foodborne illnesses, which can be caused by bacteria such as Shigella, Campylobacter, and Salmonella. Pathogenic bacteria also cause infections such as tetanus, typhoid fever, diphtheria, syphilis, and Hansen's disease. They typically range between 1 and 5 micrometers in length.",
"title": "cord19-11.txt"
},
"id": 11
}
]
Below are some explanations related to the structure of the JSON export file. The export represents a list/array of task, containing completions and/or predictions. Each task in the list has the following main elements:
TASK:
A task can have 0 or several completions and 0 or several predictions.
{
"completions": [COMPLETION1, COMPLETION2, ...],
"predictions": [PREDICTION1, PREDICTION2, ...],
"created_at": "2022-07-04 06:17:26",
"created_by": "admin",
"data": {
"text": <sample_text>"
},
"id": 1
}
completions: list of completions (manual annotations)predictions: list of predictions (annotations generated by spark-nlp model(s))created_by: time stamp representing the creation time for a taskcreated_by: the user who created the taskdata: input data on which the annotations/preannotation are defined (text/image/audio etc)id: task ID
COMPLETION/PREDICTION:
Completions and predictions have the following structure:
{
"created_username": "collaborate",
"created_ago": "2022-07-04T06:18:39.720155Z",
"lead_time": 11,
"result": [RESULT1, RESULT2, RESULT3, ....],
"honeypot": true,
"id": 1001,
"updated_at": "2022-07-04T06:18:49.037150Z",
"updated_by": "collaborate",
}
created_username: user who created the annotationcreated_ago: timestamp of when the annotation was createdlead_time: time taken (in seconds) to create this annotation (valid for manual completion only)result: list of annotated labelshoneypot: boolean value to set/unset ground truthid: completion/prediction IDupdated_at: timestamp of when the annotation was last updatedupdated_by: user who updated the annotation
Each completion/prediction contains one or several results which can be seen as individual annotations.
RESULT:
The structure of the RESULT dictionary differs according to the project configuration:
1. NER:
{
"value": {
"start": 17,
"end": 25,
"text": "pleasant",
"labels": [
"FAC"
],
"confidence": 1
},
"id": "iJOo_XgIao",
"from_name": "label",
"to_name": "text",
"type": "labels"
}
value:start: start index of the annotated chunkend: end index of the annotated chunktext: annotated chunklabels: associated labelconfidence: confidence score (1 for manual annotation and a value between 0 and 1 for predicted annotations)
id: id of annotation (used while creating relations between entities)-
from_name/to_name: this attribute is set according to the project config:from_name-> name attribute of the Labels tagto_name-> toName attribute of the Labels tag
<Labels name="label" toName="text">
type: type of the annotation (labels, choices, relations etc.)
2. Classification:
{
"value": {
"choices": [
"sadness"
],
"confidence": 1
},
"id": "VyY-OHe_lf",
"from_name": "surprise",
"to_name": "text",
"type": "choices"
}
value:choices: the options/choises selected by usersconfidence: confidence score (1 for manual annotation and between 0 and 1 for predicted annotation)
id: id of annotation-
from_name/to_name: this field is set according to the project config:`from_name` -> name attribute of the Choice tag `to_name` -> toName attribute of the Choice tag
<Choices name="surprise" toName="text" choice="single">
type: type of the annotation (labels, choices, relations etc)
3. Relations:
The information below is added in the RESULT section:
{
"from_id": "ucfP3c4xWg",
"to_id": "IlWac4TdFx",
"type": "relation",
"direction": "right",
"confidence": 1
}
from_id/to_id: IDs of the related annotationstype: type of the annotation (labels, choices, relations etc)direction: direction for the relation. The accepted values are right and left.
Submitted annotation:
When a completion is submitted, it has a submitted timestamp on the COMPLETION dictionary (refer to the above json example):
"submitted_at": "2022-07-04T12:03:48.824"
Reviewed annotation:
When a completion is reviewed, the following information is added to the COMPLETION dictionary:
"review_status": {
"approved": true,
"comment": "Looks good!",
"reviewer": "Mauro",
"reviewed_at": "2022-07-04T06:19:31.897Z"
}
approved: boolean value (true -> approved and false -> rejected)comment: text comment manually defined by the reviewerreviewer: user who reviewed the completionreviewed_at: review timestamp
Copied Annotation:
When an annotation is copied/cloned from a specific completion/prediction, the COMPLETION dictionary contains the copied_from filed:
"copied_from": "prediction: 4001"
CSV
Results are stored in a comma-separated tabular file with column names specified by “from_name” and “to_name” values.
TSV
Results are stored in a tab-separated tabular file with column names specified by “from_name” and “to_name” values.
CoNLL2003
The CoNLL export feature generates a single output file, containing all available completions for all the tasks in the project. The resulting file has the following format:
-DOCSTART- -X- O
Sample -X- _ O
Type -X- _ O
Medical -X- _ O
Specialty: -X- _ O
Endocrinology -X- _ O
Sample -X- _ O
Name: -X- _ O
Diabetes -X- _ B-Diagnosis
Mellitus -X- _ I-Diagnosis
Followup -X- _ O
Description: -X- _ O
Return -X- _ O
visit -X- _ O
to -X- _ O
the -X- _ O
endocrine -X- _ O
clinic -X- _ O
for -X- _ O
followup -X- _ O
management -X- _ O
of -X- _ O
type -X- _ O
1 -X- _ O
diabetes -X- _ O
mellitus -X- _ O
Plan -X- _ O
today -X- _ O
is -X- _ O
to -X- _ O
make -X- _ O
adjustments -X- _ O
to -X- _ O
her -X- _ O
pump -X- _ O
based -X- _ O
on -X- _ O
a -X- _ O
total -X- _ O
daily -X- _ B-FREQUENCY
dose -X- _ O
of -X- _ O
90 -X- _ O
units -X- _ O
of -X- _ O
insulin -X- _ O
…
Users can specify if only starred completions should be included in the output file by checking the Only ground truth option before generating the export.
Supported Formats for Visual NER Projects
The process of annotations export from Visual NER projects is similar to that of text projects.
When exporting the Visual NER annotations users have two additional formats available:
COCO
The COCO format is a specific JSON structure dictating how labels and metadata are saved for an image dataset. It is a large-scale object detection, segmentation, and captioning dataset. Exporting in COCO format is available for Visual NER projects only.
Below is a sample format:
{
"images": [
{
"width": 6.588235294117647,
"height": 0.9396786905122766,
"id": 0,
"file_name": [
"/images/19/0160023239a-1655481445_0.png",
"/images/19/0160023239a-1655481445_1.png"
]
}
],
"categories": [
{
"id": 0,
"name": "OGSContractNumber",
"supercategory": "OGSContractNumber"
},
{
"id": 1,
"name": "Contractor",
"supercategory": "Contractor"
},
{
"id": 2,
"name": "FederalID",
"supercategory": "FederalID"
},
{
"id": 3,
"name": "VendorID",
"supercategory": "VendorID"
},
{
"id": 4,
"name": "Title",
"supercategory": "Title"
},
{
"id": 5,
"name": "AwardNumber",
"supercategory": "AwardNumber"
},
{
"id": 6,
"name": "ContractPeriod",
"supercategory": "ContractPeriod"
},
{
"id": 7,
"name": "BidOpeningDate",
"supercategory": "BidOpeningDate"
},
{
"id": 8,
"name": "DateOfIssue",
"supercategory": "DateOfIssue"
},
{
"id": 9,
"name": "SpecificationReference",
"supercategory": "SpecificationReference"
},
{
"id": 10,
"name": "GroupNumber",
"supercategory": "GroupNumber"
}
],
"annotations": [
{
"id": 0,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69434",
"pageNumber": 2
},
{
"id": 1,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69435",
"pageNumber": 2
},
{
"id": 2,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69436",
"pageNumber": 2
},
{
"id": 3,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
1,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Cream-O-Land Dairies, LLC",
"pageNumber": 2
},
{
"id": 4,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
1,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Hudson Valley Fresh Dairy, LLC",
"pageNumber": 2
},
{
"id": 5,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
1,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Upstate Niagara Inc.",
"pageNumber": 2
},
{
"id": 6,
"image_id": 0,
"category_id": 2,
"segmentation": [],
"bbox": [
3,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "223629742",
"pageNumber": 2
},
{
"id": 7,
"image_id": 0,
"category_id": 2,
"segmentation": [],
"bbox": [
3,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "461053272",
"pageNumber": 2
},
{
"id": 8,
"image_id": 0,
"category_id": 2,
"segmentation": [],
"bbox": [
3,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "160845625",
"pageNumber": 2
},
{
"id": 9,
"image_id": 0,
"category_id": 3,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "1100070111",
"pageNumber": 2
},
{
"id": 10,
"image_id": 0,
"category_id": 3,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "1100212977",
"pageNumber": 2
},
{
"id": 11,
"image_id": 0,
"category_id": 3,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "1000014941",
"pageNumber": 2
},
{
"id": 12,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
4,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Cream-O-Land - LLC",
"pageNumber": 2
},
{
"id": 13,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69434",
"pageNumber": 2
},
{
"id": 14,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
4,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Hudson Valley Fresh Dairy, LLC",
"pageNumber": 2
},
{
"id": 15,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69435",
"pageNumber": 2
},
{
"id": 16,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69436",
"pageNumber": 2
},
{
"id": 17,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
4,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Upstate Niagara Cooperative, Inc.",
"pageNumber": 2
},
{
"id": 18,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
4,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "; Upstate Niagara Cooperative,",
"pageNumber": 2
},
{
"id": 19,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69436",
"pageNumber": 2
},
{
"id": 20,
"image_id": 0,
"category_id": 4,
"segmentation": [],
"bbox": [
3,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Milk, Fluid (Statewide)",
"pageNumber": 1
},
{
"id": 21,
"image_id": 0,
"category_id": 5,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "23239",
"pageNumber": 1
},
{
"id": 22,
"image_id": 0,
"category_id": 6,
"segmentation": [],
"bbox": [
2,
0,
2,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "September 21, 2021 Through September 20, 2026",
"pageNumber": 1
},
{
"id": 23,
"image_id": 0,
"category_id": 7,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "June 10, 2021",
"pageNumber": 1
},
{
"id": 24,
"image_id": 0,
"category_id": 8,
"segmentation": [],
"bbox": [
2,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "September 14, 2021",
"pageNumber": 1
},
{
"id": 27,
"image_id": 0,
"category_id": 10,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Group",
"pageNumber": 1
},
{
"id": 29,
"image_id": 0,
"category_id": 4,
"segmentation": [],
"bbox": [
3,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Milk, Fluid (Statewide)",
"pageNumber": 1
},
{
"id": 30,
"image_id": 0,
"category_id": 5,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "23239",
"pageNumber": 1
},
{
"id": 31,
"image_id": 0,
"category_id": 6,
"segmentation": [],
"bbox": [
2,
0,
2,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "September 21, 2021 Through September 20, 2026",
"pageNumber": 1
},
{
"id": 32,
"image_id": 0,
"category_id": 6,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "June 10, 2021",
"pageNumber": 1
},
{
"id": 33,
"image_id": 0,
"category_id": 6,
"segmentation": [],
"bbox": [
2,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "September 14, 2021",
"pageNumber": 1
},
{
"id": 34,
"image_id": 0,
"category_id": 5,
"segmentation": [],
"bbox": [
2,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "23239",
"pageNumber": 1
},
{
"id": 35,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69434",
"pageNumber": 2
},
{
"id": 36,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
1,
0,
1,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Cream-O-Land Dairies, LLC",
"pageNumber": 2
},
{
"id": 38,
"image_id": 0,
"category_id": 3,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "1100070111",
"pageNumber": 2
},
{
"id": 39,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69435",
"pageNumber": 2
},
{
"id": 41,
"image_id": 0,
"category_id": 2,
"segmentation": [],
"bbox": [
3,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "461053272",
"pageNumber": 2
},
{
"id": 43,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
0,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69436",
"pageNumber": 2
},
{
"id": 45,
"image_id": 0,
"category_id": 2,
"segmentation": [],
"bbox": [
3,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "160845625",
"pageNumber": 2
},
{
"id": 47,
"image_id": 0,
"category_id": 1,
"segmentation": [],
"bbox": [
4,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "Cream-O-Land",
"pageNumber": 2
},
{
"id": 49,
"image_id": 0,
"category_id": 0,
"segmentation": [],
"bbox": [
5,
0,
0,
0
],
"ignore": 0,
"iscrowd": 0,
"area": 0,
"text": "PC69434",
"pageNumber": 2
}
],
"info": {
"year": 2022,
"version": "1.0",
"contributor": "Generative AI Lab Converter"
}
}
Pascal VOC XML
Pascal Visual Object Classes(VOC) is an XML file that contains the image details, bounding box details, classes, pose, truncated, and other data. For each image of the task there will be an XML annotation file. Exporting in VOC format is available for Visual NER projects only.
Below is a sample format:
<?xml version="1.0" encoding="utf-8"?>
<annotation>
<folder>images</folder>
<filename>0160023239a-1655481445_0.png</filename>
<source>
<database>ALABDB</database>
</source>
<owner>
<name>AnnotationLab</name>
</owner>
<size>
<width>2550</width>
<height>3299</height>
<depth>1</depth>
</size>
<segmented>0</segmented>
<object>
<name>Title</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>1305</xmin>
<ymin>660</ymin>
<xmax>1780</xmax>
<ymax>703</ymax>
</bndbox>
</object>
<object>
<name>AwardNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>973</xmin>
<ymin>791</ymin>
<xmax>1099</xmax>
<ymax>834</ymax>
</bndbox>
</object>
<object>
<name>ContractPeriod</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>903</ymin>
<xmax>2038</xmax>
<ymax>946</ymax>
</bndbox>
</object>
<object>
<name>BidOpeningDate</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>1013</ymin>
<xmax>1263</xmax>
<ymax>1054</ymax>
</bndbox>
</object>
<object>
<name>DateOfIssue</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>1124</ymin>
<xmax>1393</xmax>
<ymax>1166</ymax>
</bndbox>
</object>
<object>
<name>SpecificationReference</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>973</xmin>
<ymin>1235</ymin>
<xmax>1729</xmax>
<ymax>1277</ymax>
</bndbox>
</object>
<object>
<name>GroupNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>660</ymin>
<xmax>1248</xmax>
<ymax>702</ymax>
</bndbox>
</object>
<object>
<name>GroupNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>660</ymin>
<xmax>1108</xmax>
<ymax>702</ymax>
</bndbox>
</object>
<object>
<name>AwardNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>1125</xmin>
<ymin>660</ymin>
<xmax>1248</xmax>
<ymax>694</ymax>
</bndbox>
</object>
<object>
<name>Title</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>1304</xmin>
<ymin>660</ymin>
<xmax>1780</xmax>
<ymax>703</ymax>
</bndbox>
</object>
<object>
<name>AwardNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>791</ymin>
<xmax>1097</xmax>
<ymax>825</ymax>
</bndbox>
</object>
<object>
<name>ContractPeriod</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>902</ymin>
<xmax>2038</xmax>
<ymax>945</ymax>
</bndbox>
</object>
<object>
<name>ContractPeriod</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>1013</ymin>
<xmax>1264</xmax>
<ymax>1054</ymax>
</bndbox>
</object>
<object>
<name>ContractPeriod</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>974</xmin>
<ymin>1124</ymin>
<xmax>1393</xmax>
<ymax>1166</ymax>
</bndbox>
</object>
<object>
<name>AwardNumber</name>
<pose>Unspecified</pose>
<truncated>0</truncated>
<difficult>0</difficult>
<bndbox>
<xmin>791</xmin>
<ymin>2246</ymin>
<xmax>903</xmax>
<ymax>2276</ymax>
</bndbox>
</object>
</annotation>
Export Options
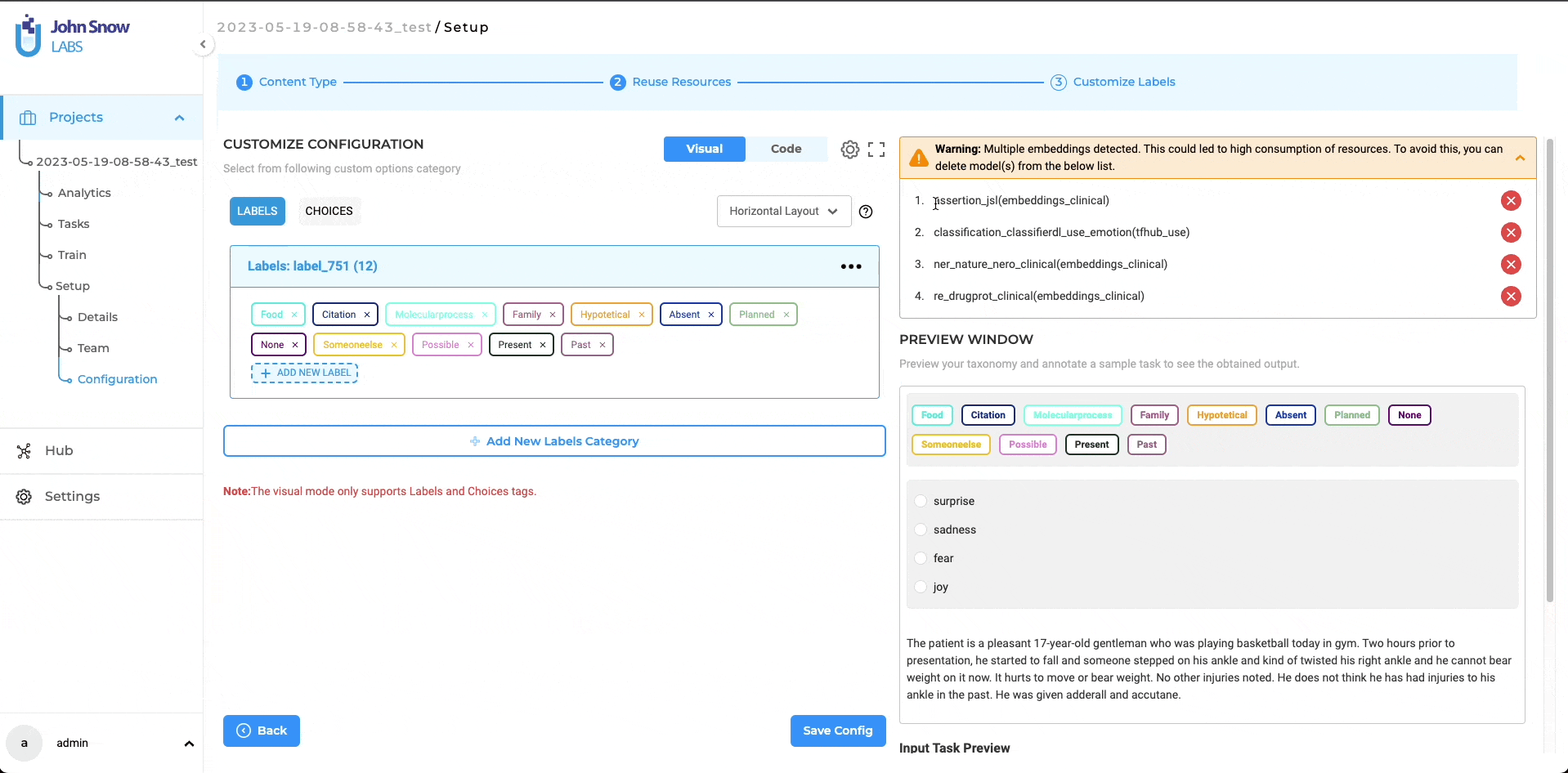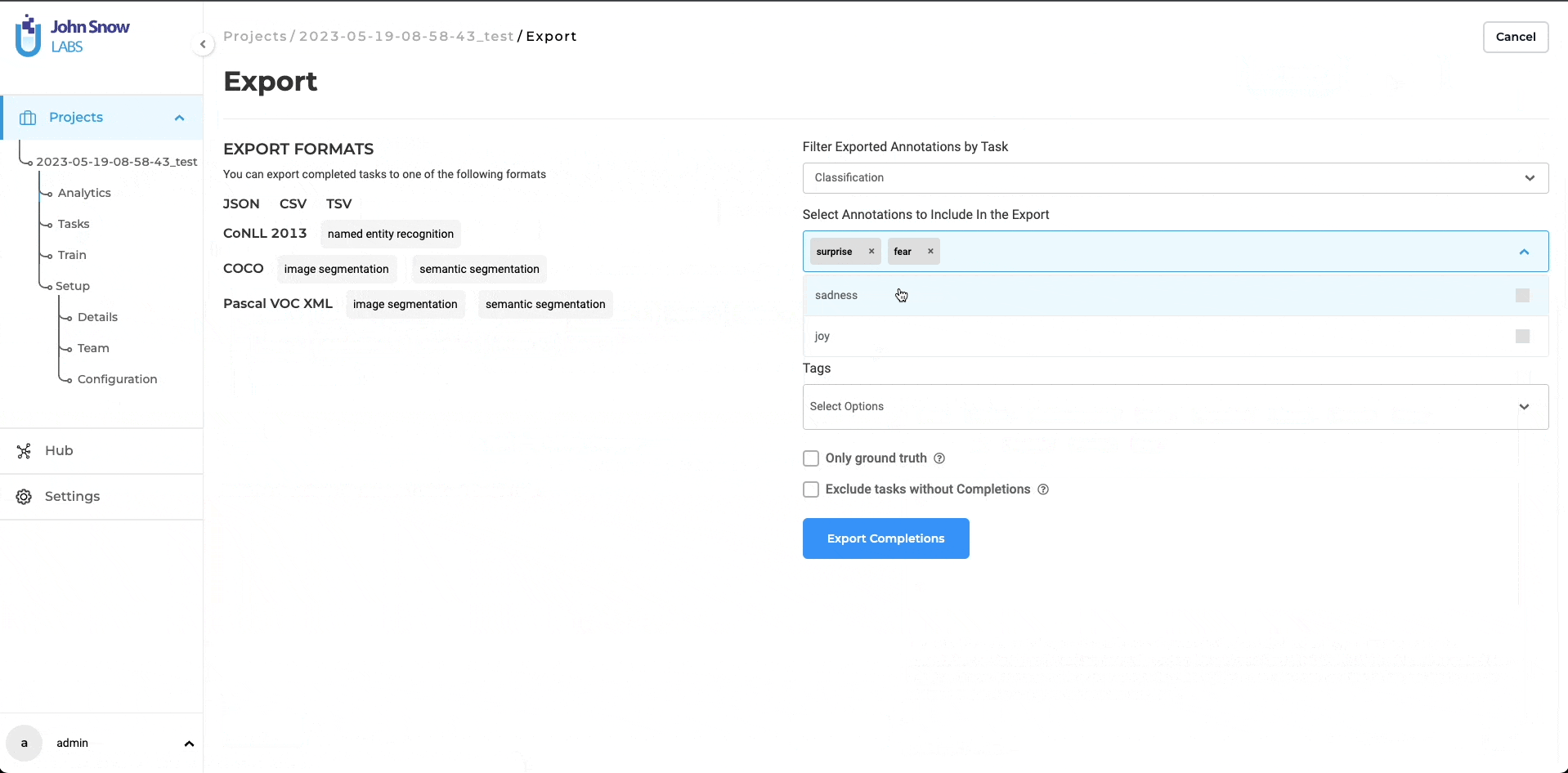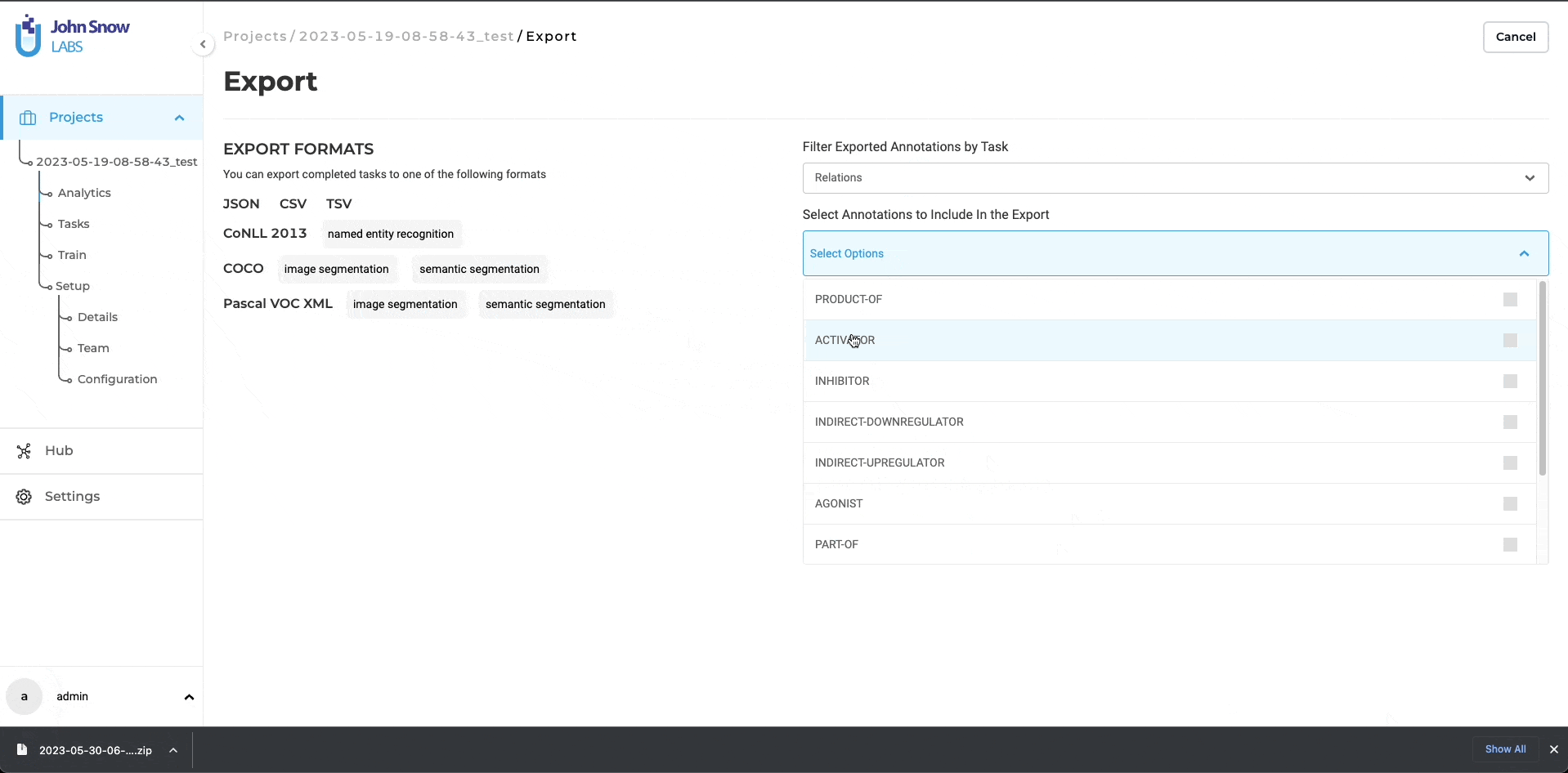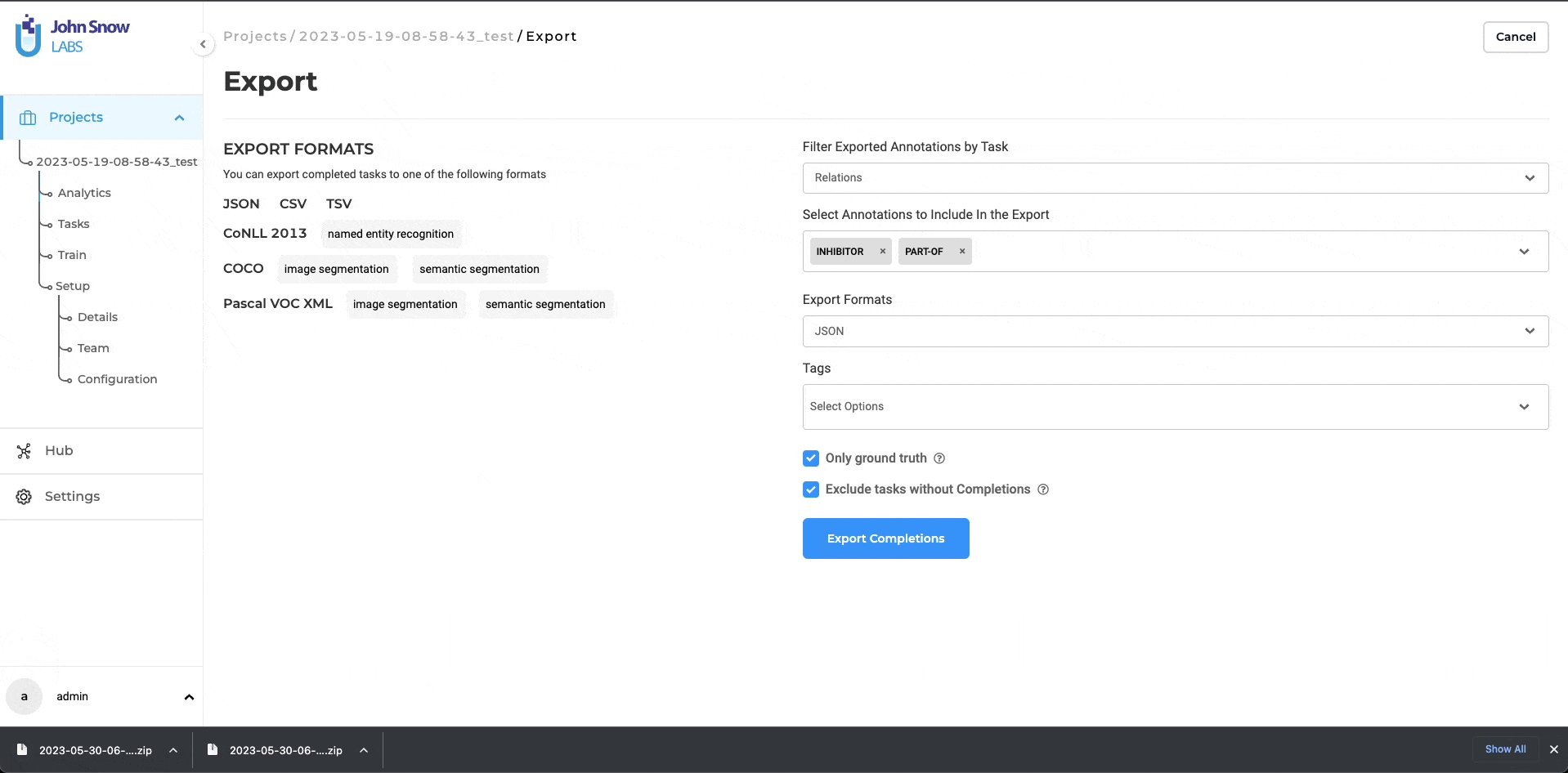
Filter Exported Annotations by Task
This filter allows users to select annotations based on the task (NER, Classification, Assertion, Relation Extraction)
Select Annotations to Include In the Export
This filter can be used to select available labels, classes, assertion labels, or relations.

Tags
Only allow export of tasks having the specified tags.
Only Ground Truth
If this option is enabled then only the tasks having ground truth in the completion will be exported.
Exclude tasks without Completions
You can choose whether to include tasks that have no completions in the export.
Enable Exclude tasks without Completions on the Export page to export only tasks that contain at least one completion (useful when you don’t want empty tasks in downstream workflows).
Leave it unchecked to include all tasks—completed and empty—such as when cloning or archiving full projects.
Integration with Amazon S3 and Azure Blob for Tasks and Projects Export
Generative AI Lab provides seamless integration with Amazon S3 and Azure Blob Storage, enabling users to export annotated tasks and entire projects directly to cloud storage.
This simplifies data management and ensures a smooth transition from annotation to model training or deployment.
When triggering an export, a popup window prompts users to select the target destination.
By default, the Local Export tab is selected, meaning files are downloaded to the local workstation.
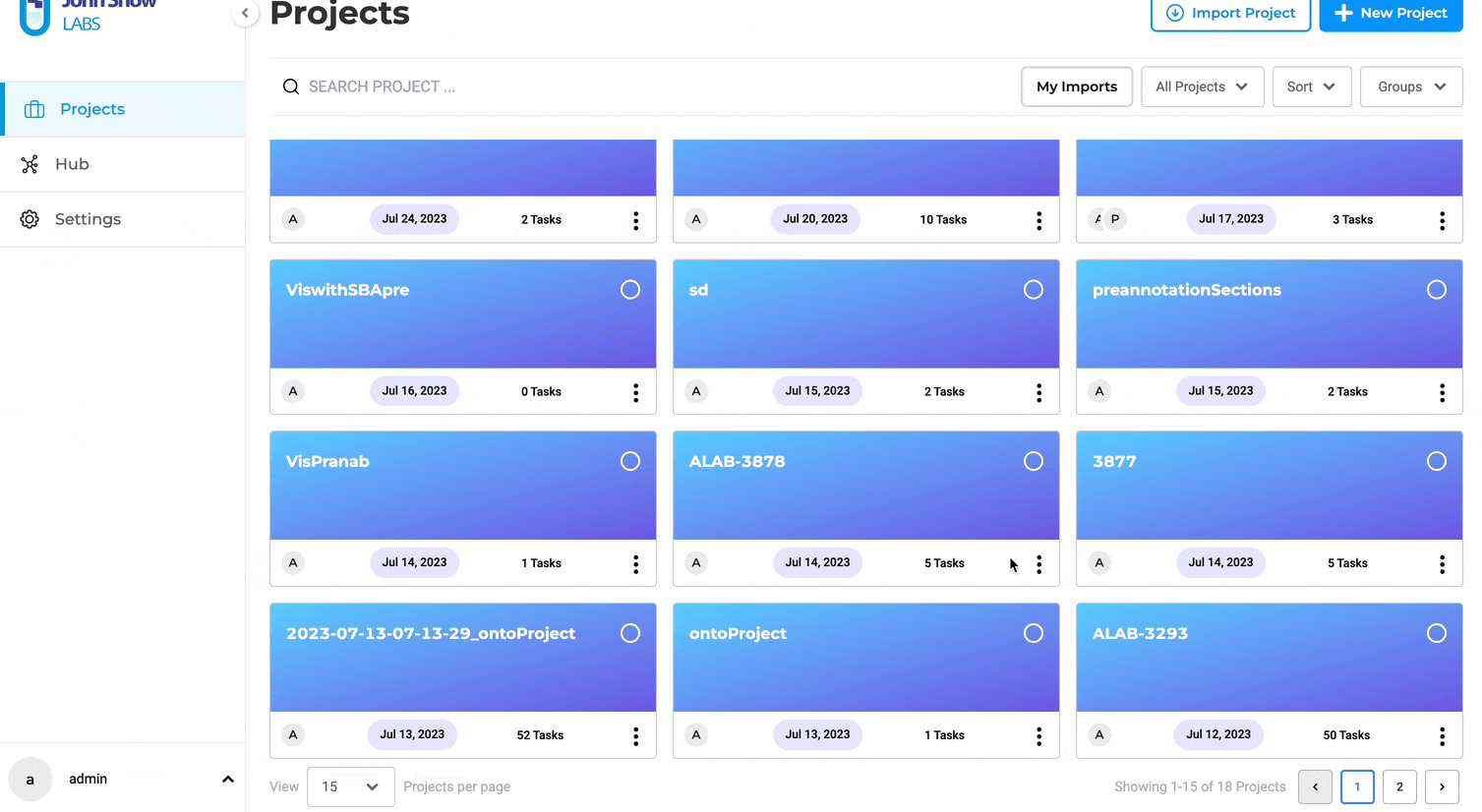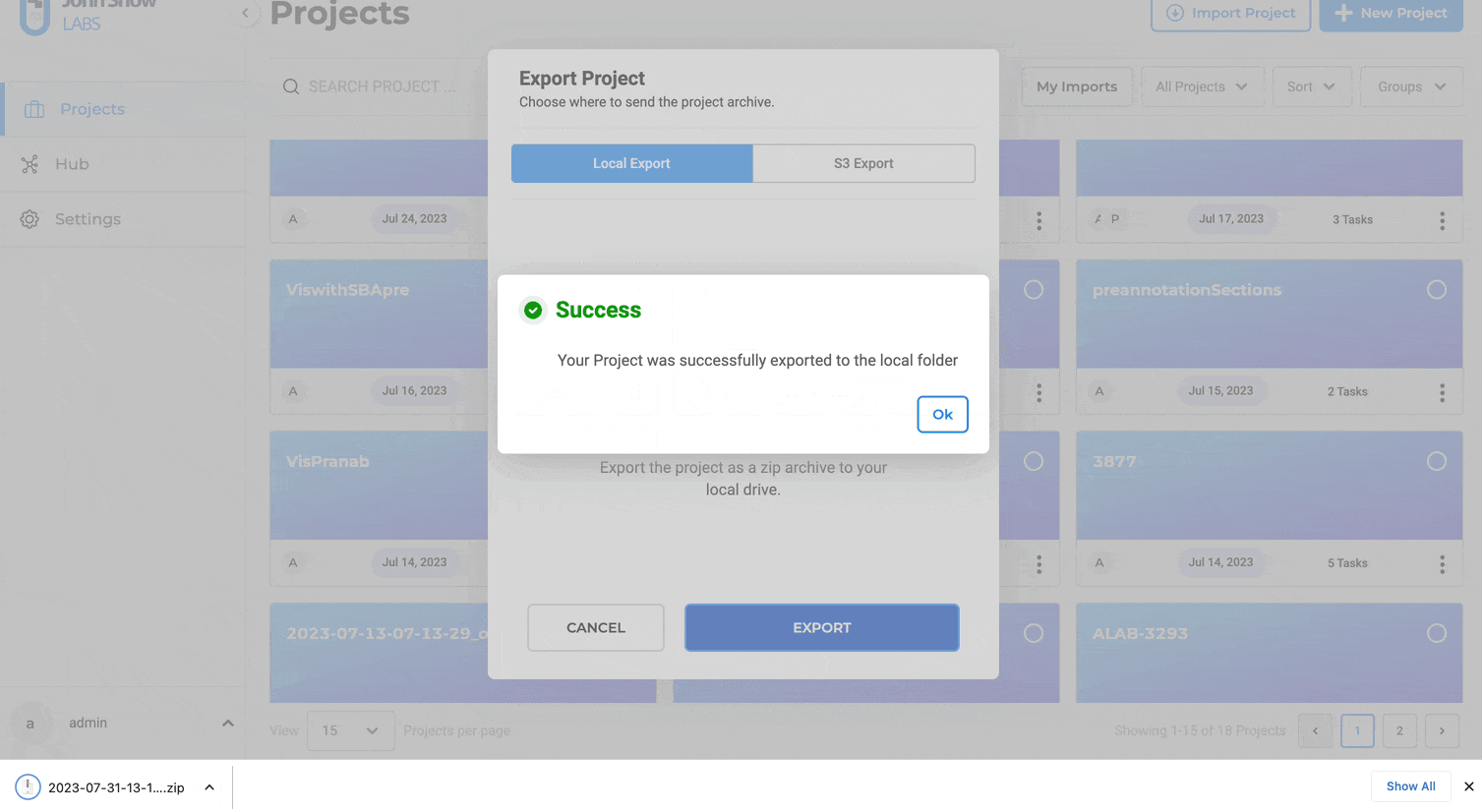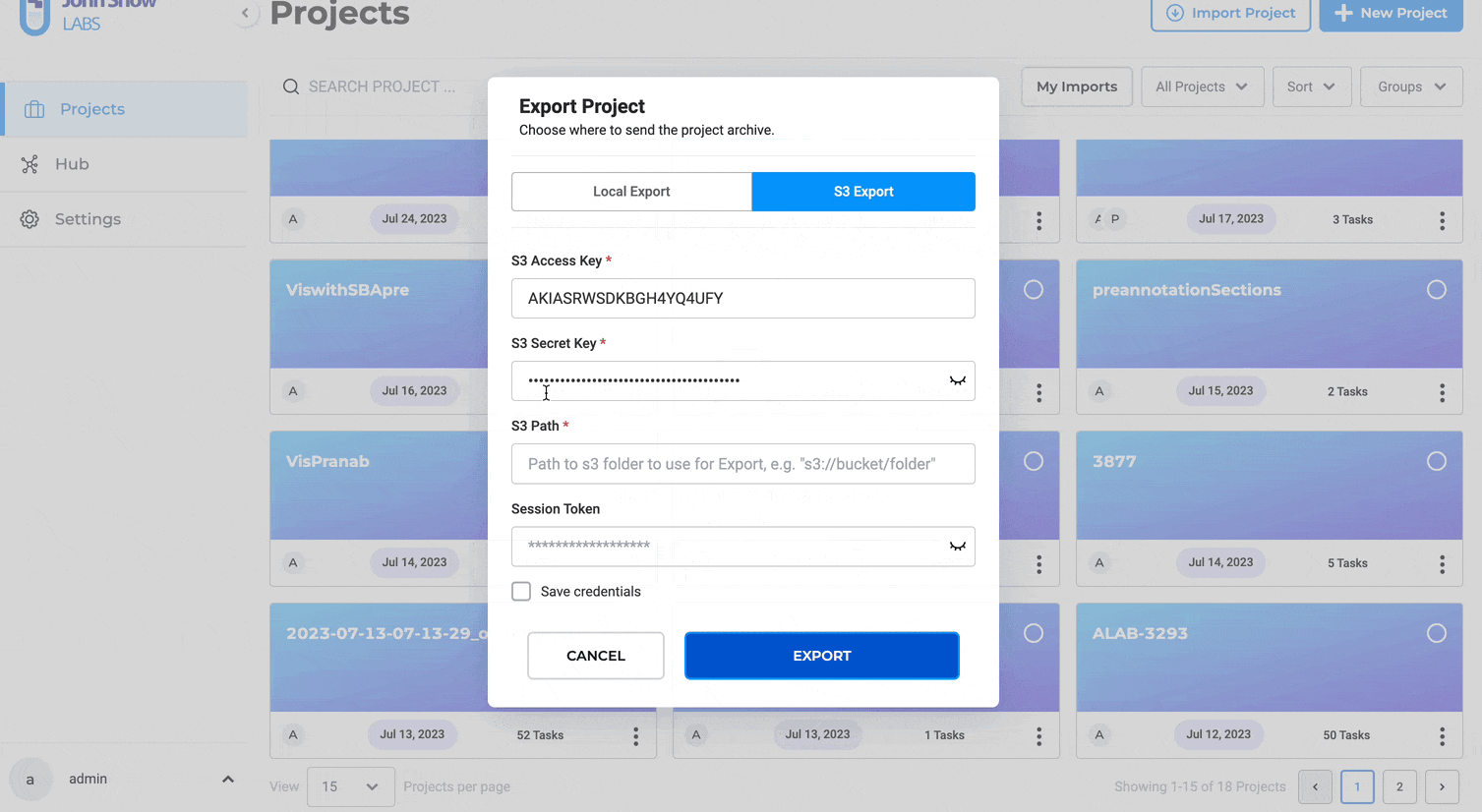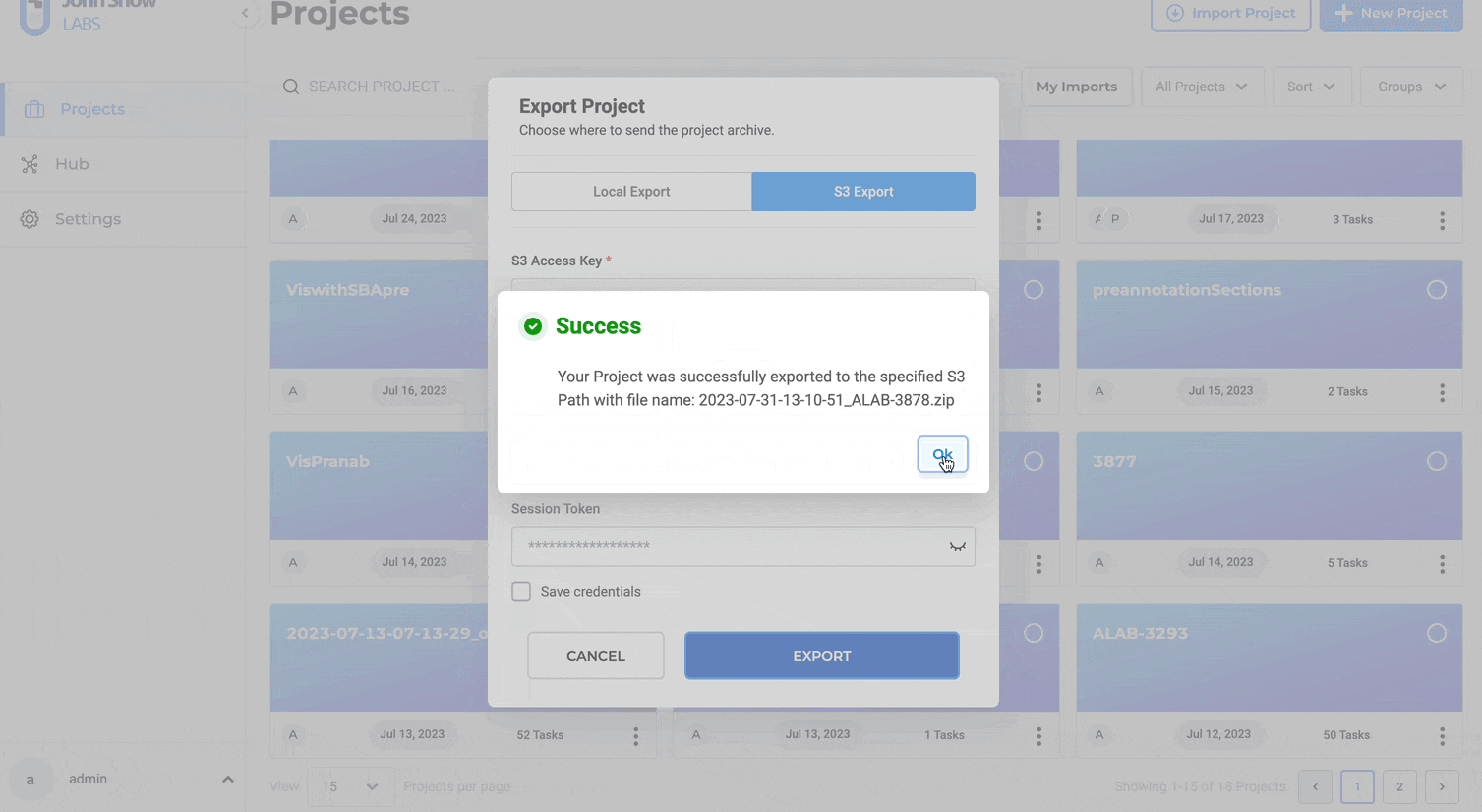
Alternatively, users can switch to the S3 Export tab to send project data directly to an Amazon S3 bucket or to the Azure Blob Export tab for Azure storage.
S3 credentials can be entered manually and securely saved for future use.
Note: When running on an AWS EC2 instance with an attached IAM role, Generative AI Lab automatically uses that role’s permissions for S3 exports—no manual AWS keys are required.
Seamless Task Export to Azure Blob Storage:
Exporting projects to Azure Blob storage is now an equally streamlined process:
- Access Export Page: Navigate to the “Export Tasks” page within the Generative AI Lab platform.
- Specify the Tasks to Export: Use the filter on the page to select the tasks you want to export as well as the target format and click the Export button.
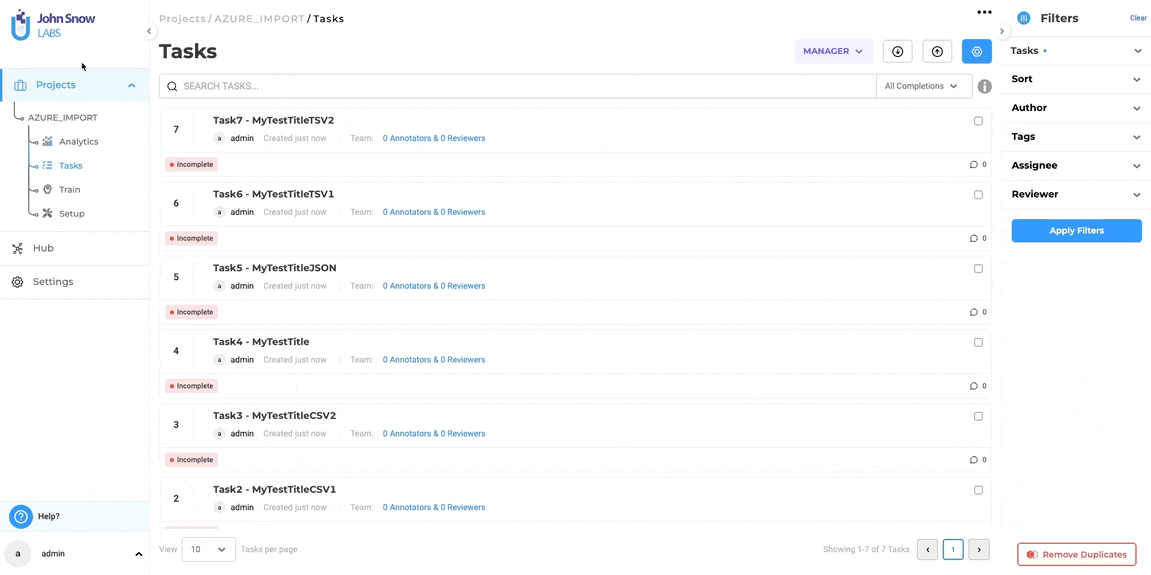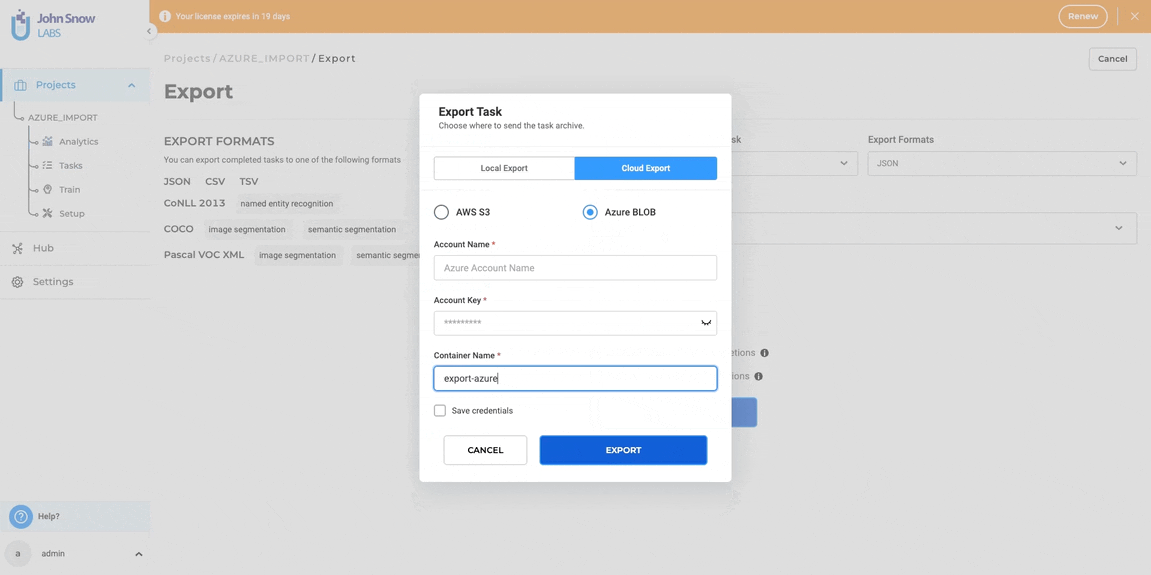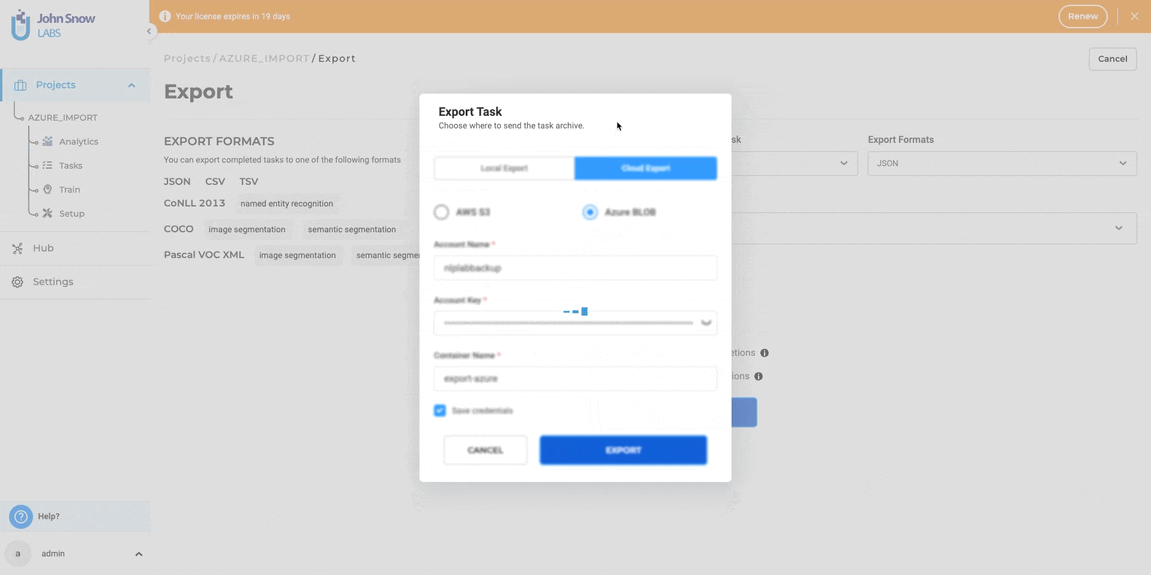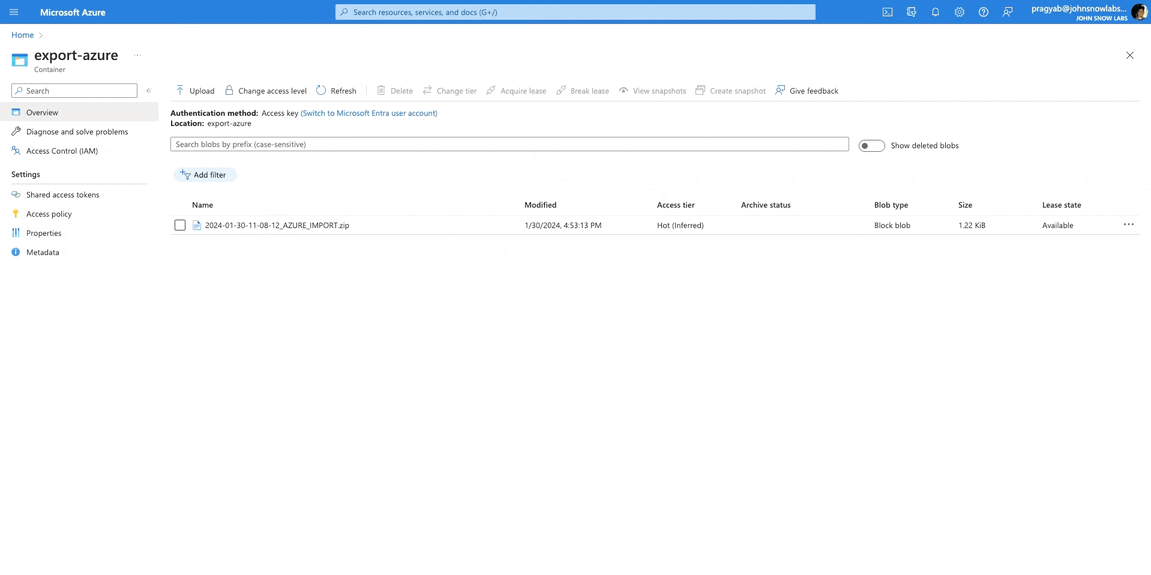
- Select Cloud Export Option: Navigate to the “Cloud Export” tab on the pop-up and select “Azure BLOB” from the available cloud storage options.
- Enter Azure Credentials: Provide the Azure connection details: Azure Container Name, Azure Account Name, and Azure Account Secret Key.
- Optionally Save Credentials: Save the credentials for future use to expedite subsequent exports to Azure Blob storage.
- Initiate Export Process: Click the “Export” button to seamlessly transfer the selected project tasks into the specified Azure Blob container, ensuring effortless data backup and management.
This integration with Azure Blob storage empowers Generative AI Lab users to manage tasks with unparalleled efficiency and flexibility. By leveraging the power of Azure, users can seamlessly import and export tasks, streamline data handling processes, and enhance their overall Generative AI Lab experience.
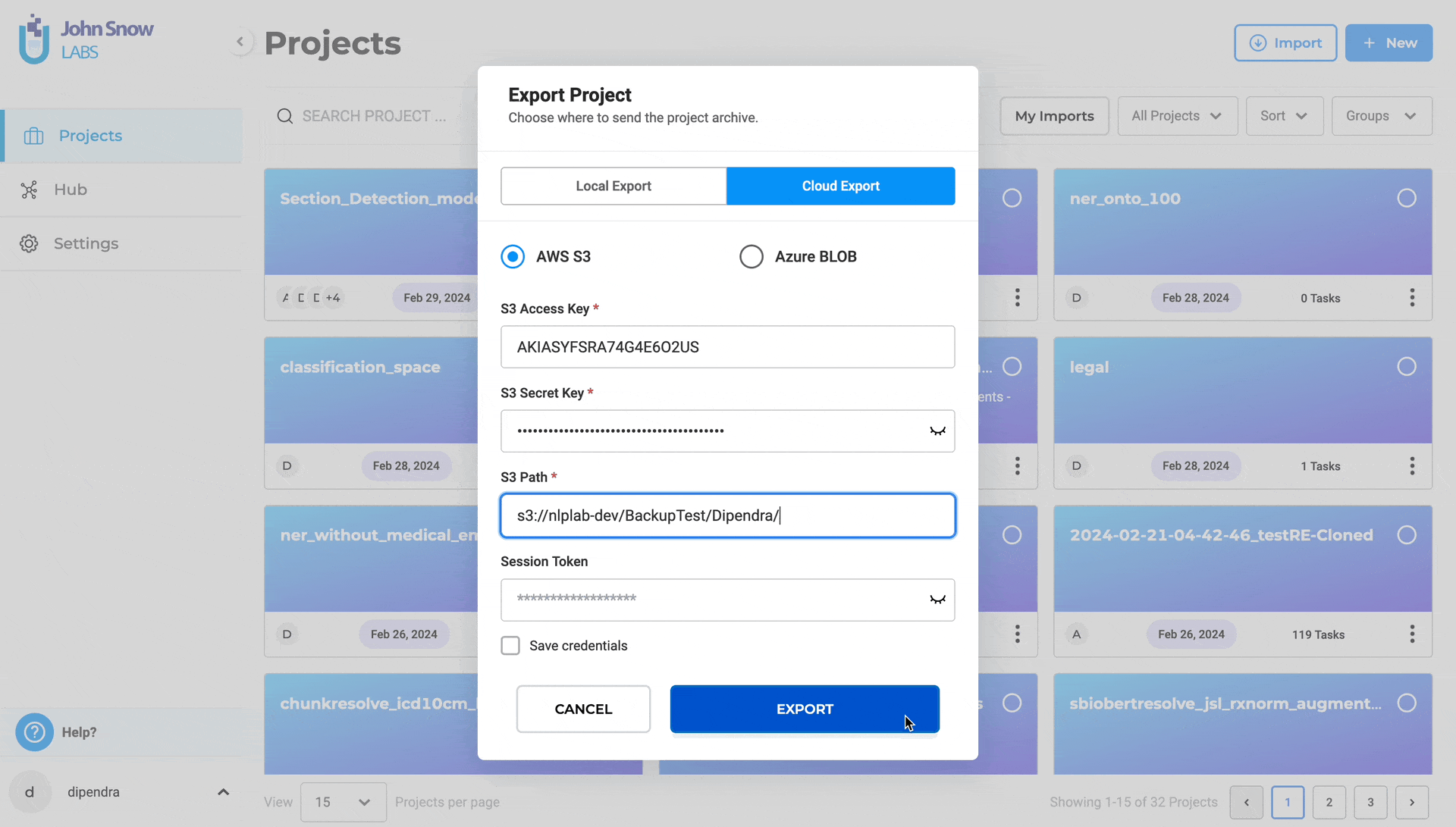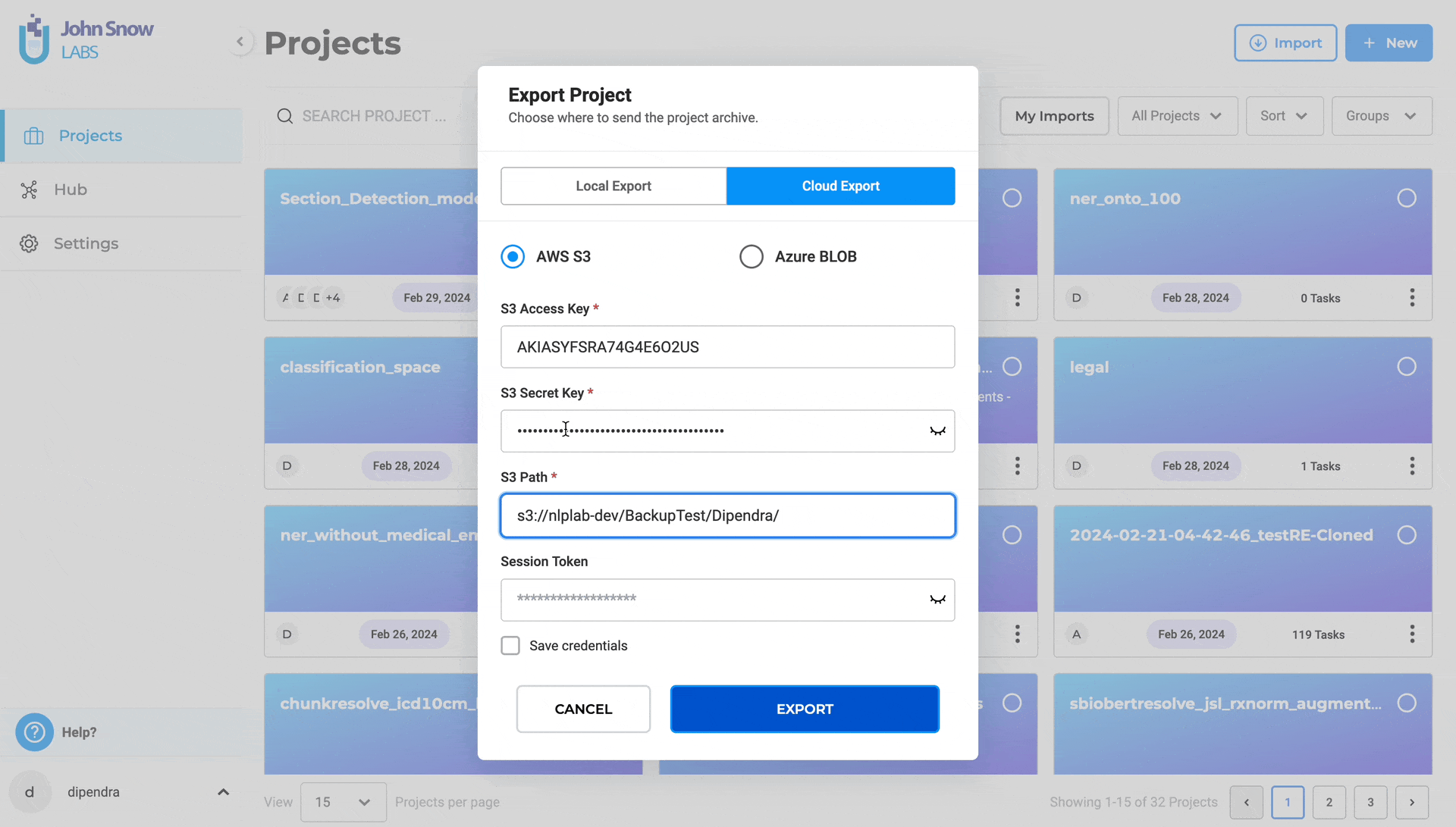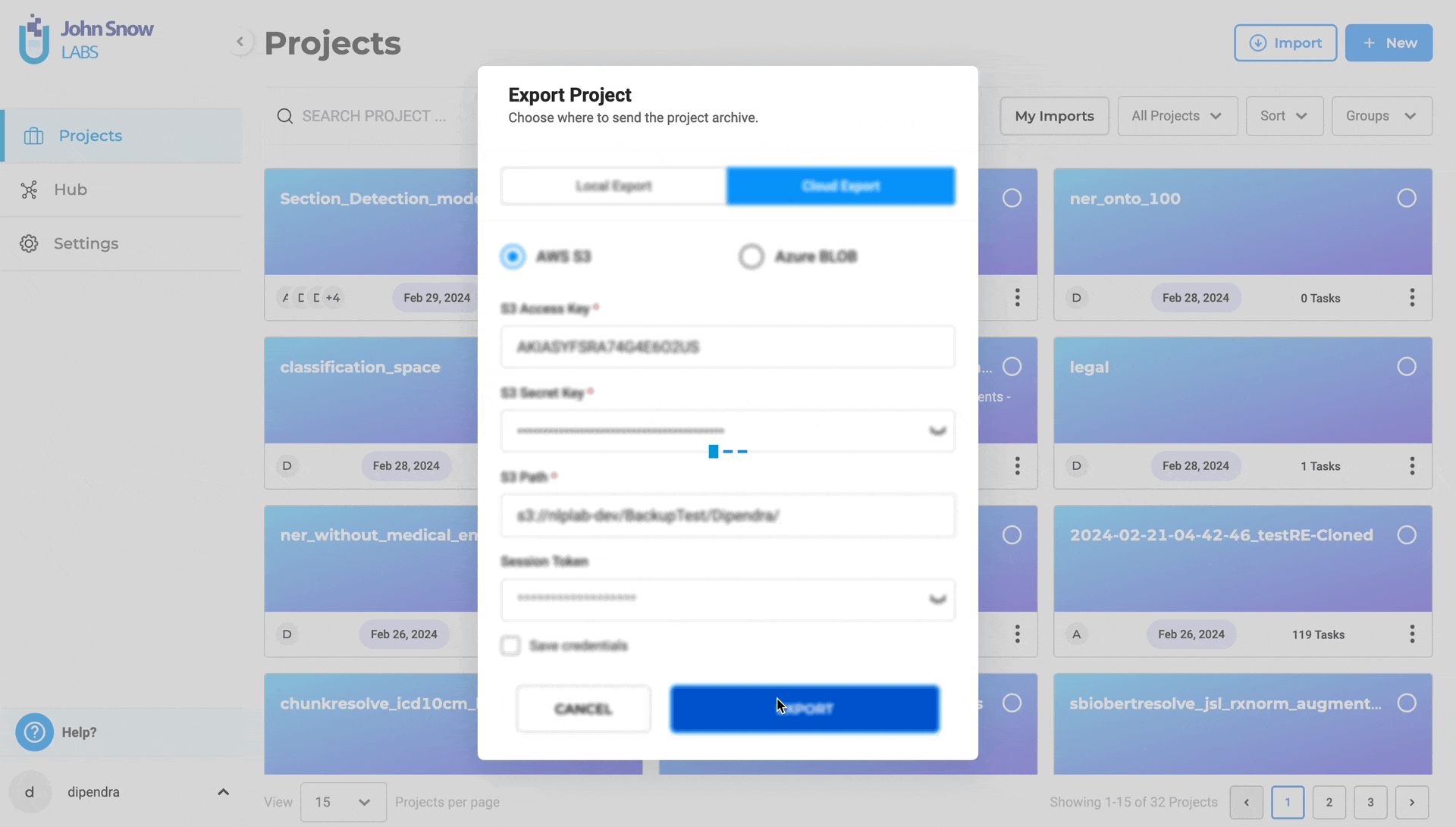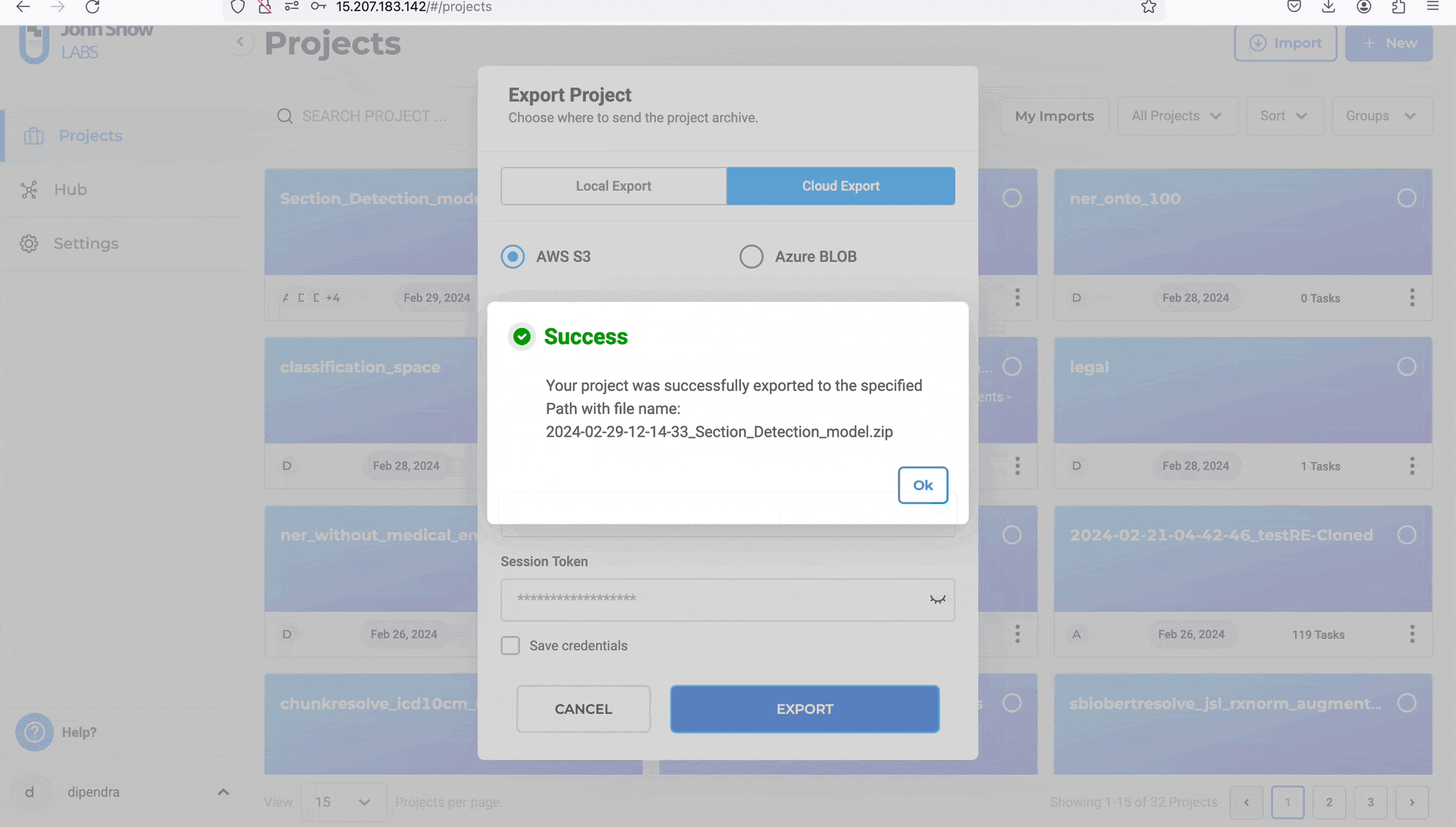
Steps to export a project to S3:
- Navigate to “Projects”
- Choose the desired project and Click “Export Project”
- Select “Cloud Export”
- Click “AWS S3”
- Input S3 Access Key and S3 Secret Key
- Specify the S3 path for export (e.g., s3://bucket/folder/)
- Optionally, provide Session Token for MFA Account
- You click on Save Credentils as well for the future use
- Optionally, save credentials for future use
- Click “EXPORT”
Note:
When exporting projects to AWS S3, if Generative AI Lab is running on an AWS EC2 instance with an attached IAM role, the platform can automatically use that role’s credentials for authentication.
In this case, you do not need to manually enter or store AWS Access or Secret Keys—the system securely uses the IAM role permissions assigned to the instance to perform the export operation.
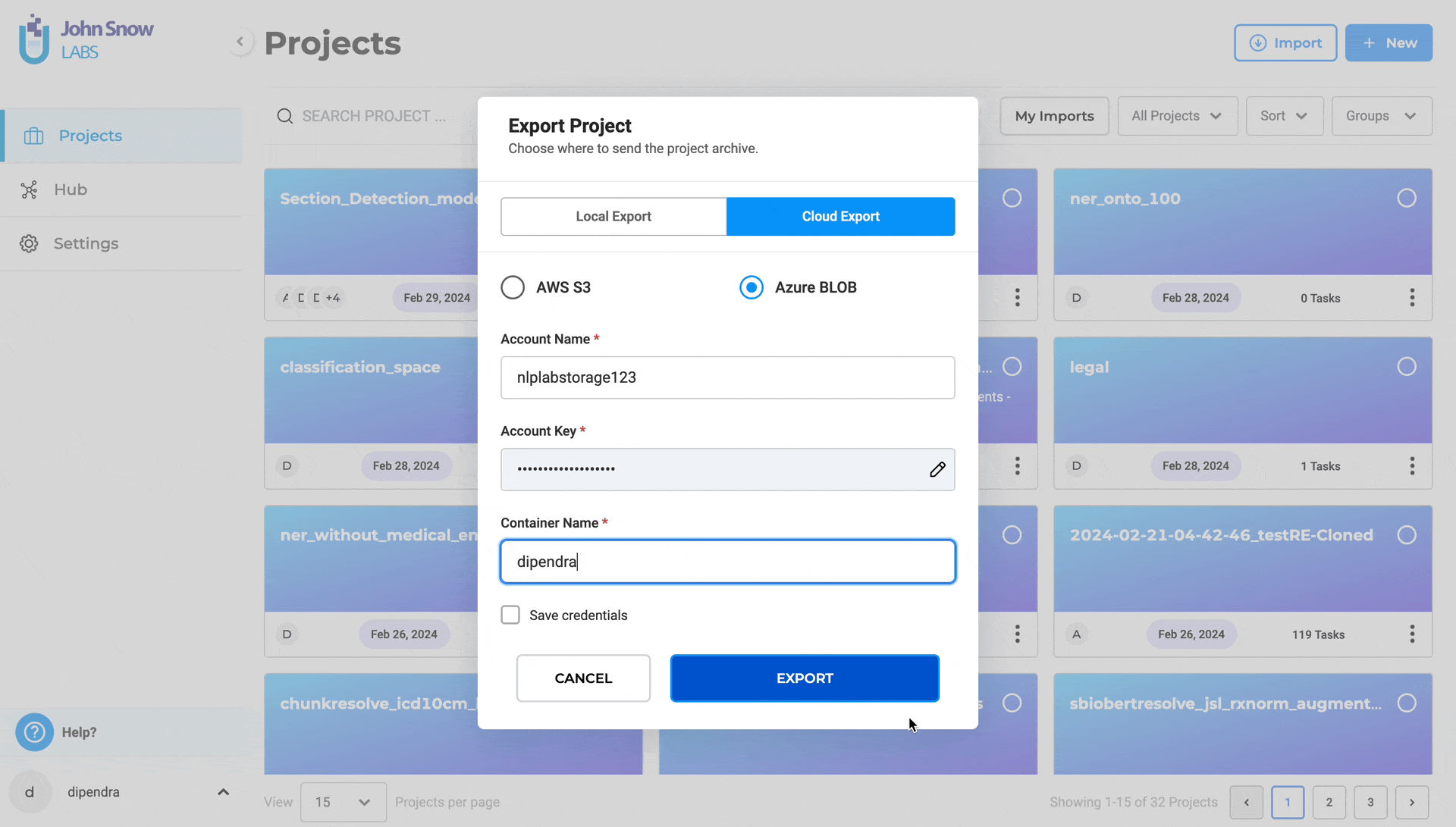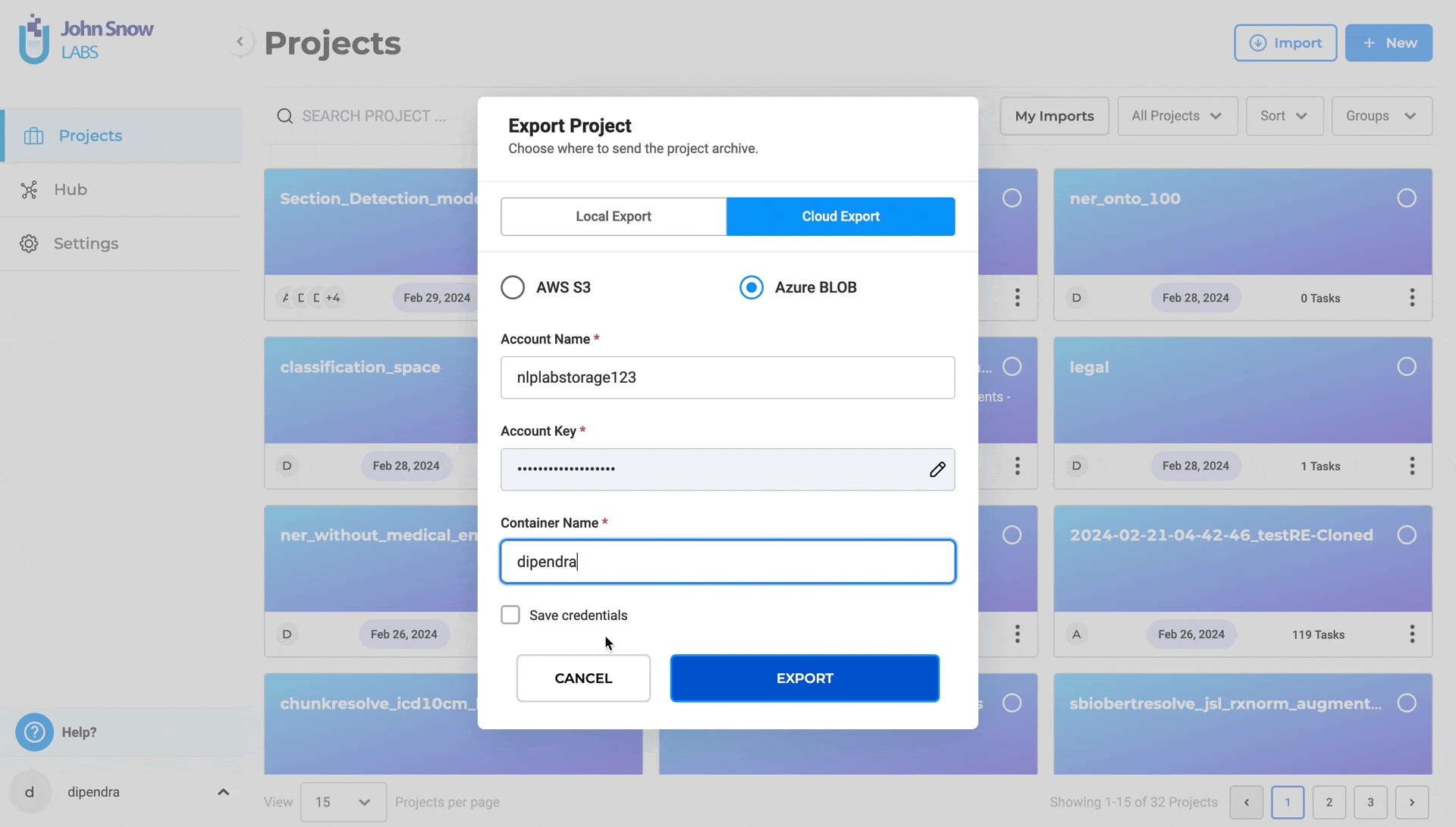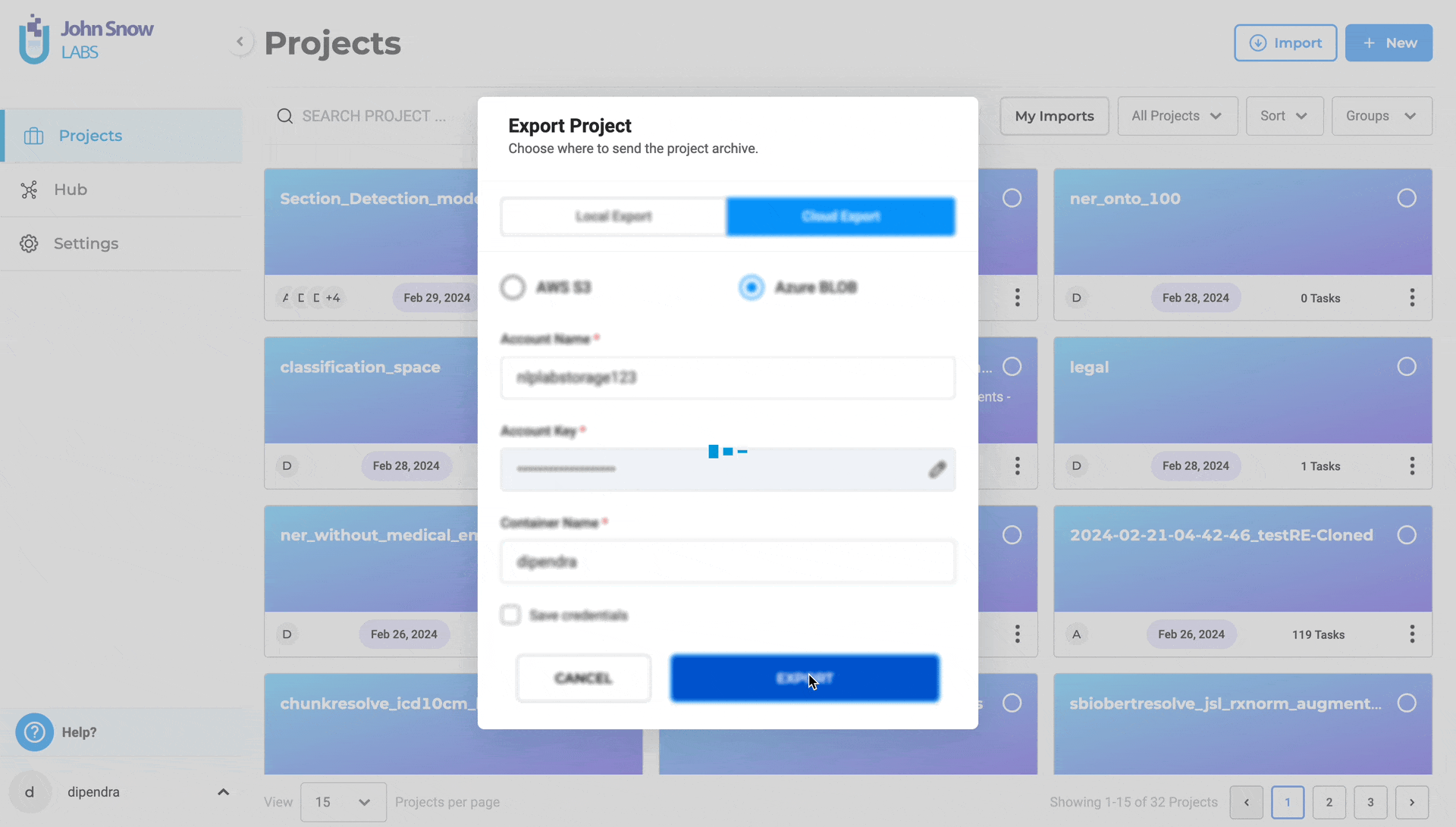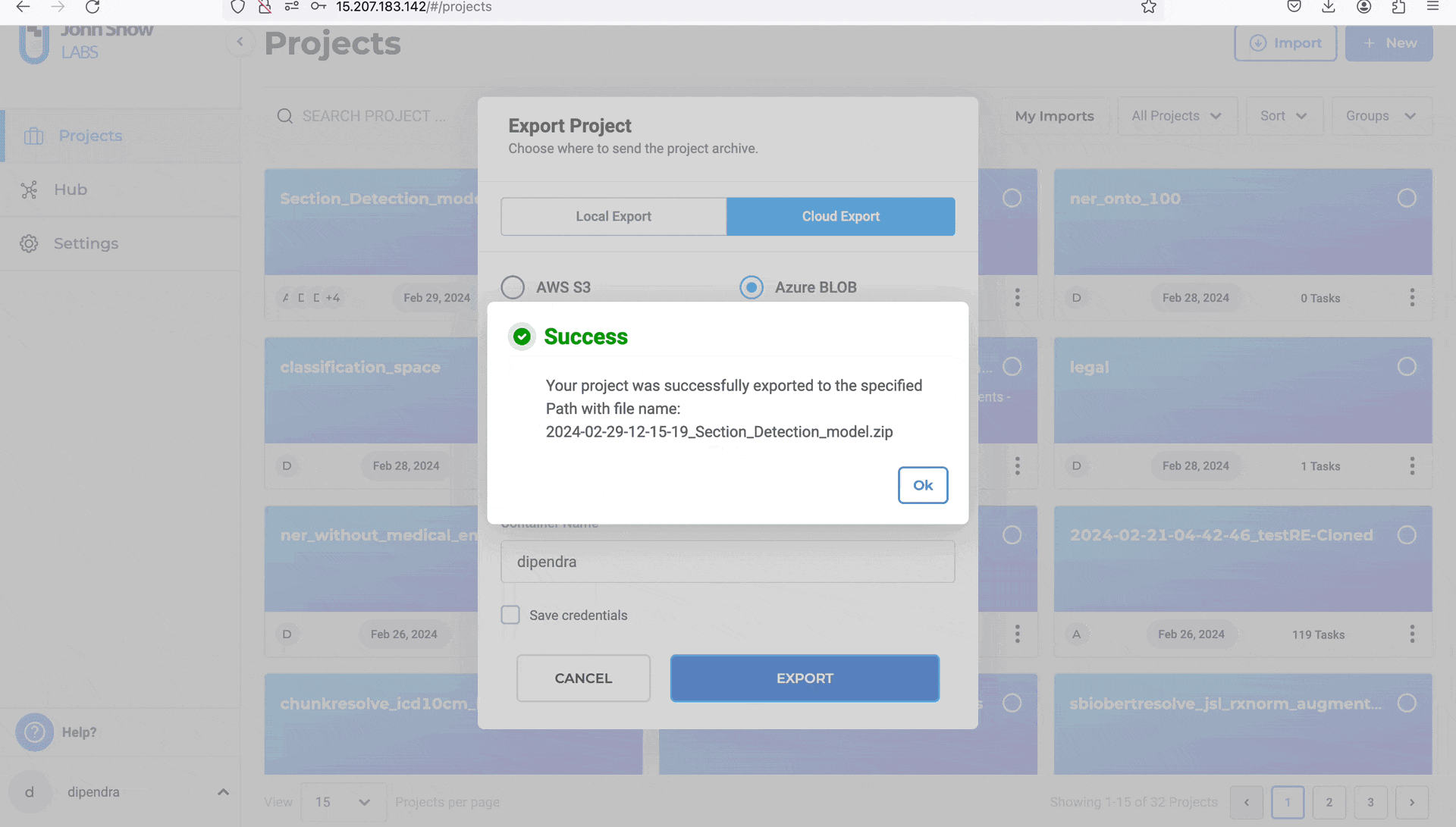
Steps to export a project to Azure Blob:
- Navigate to “Projects”
- Select the project and Click “Export Project”
- Choose “Cloud Export”
- Select “Azure Blob”
- Enter Account Name, Account Key, and Container Name
- Optionally, save credentials for future use
- Click “EXPORT”
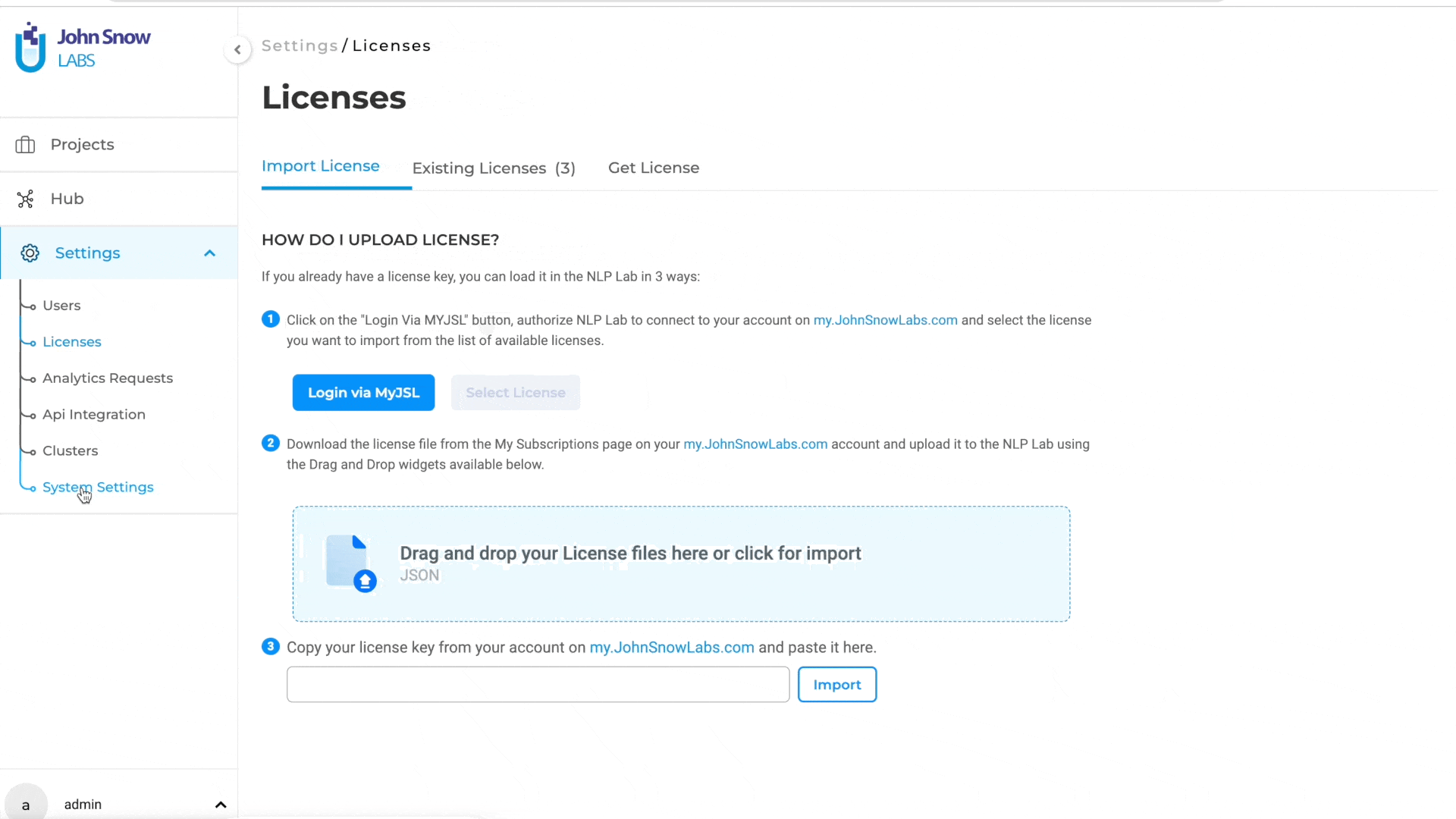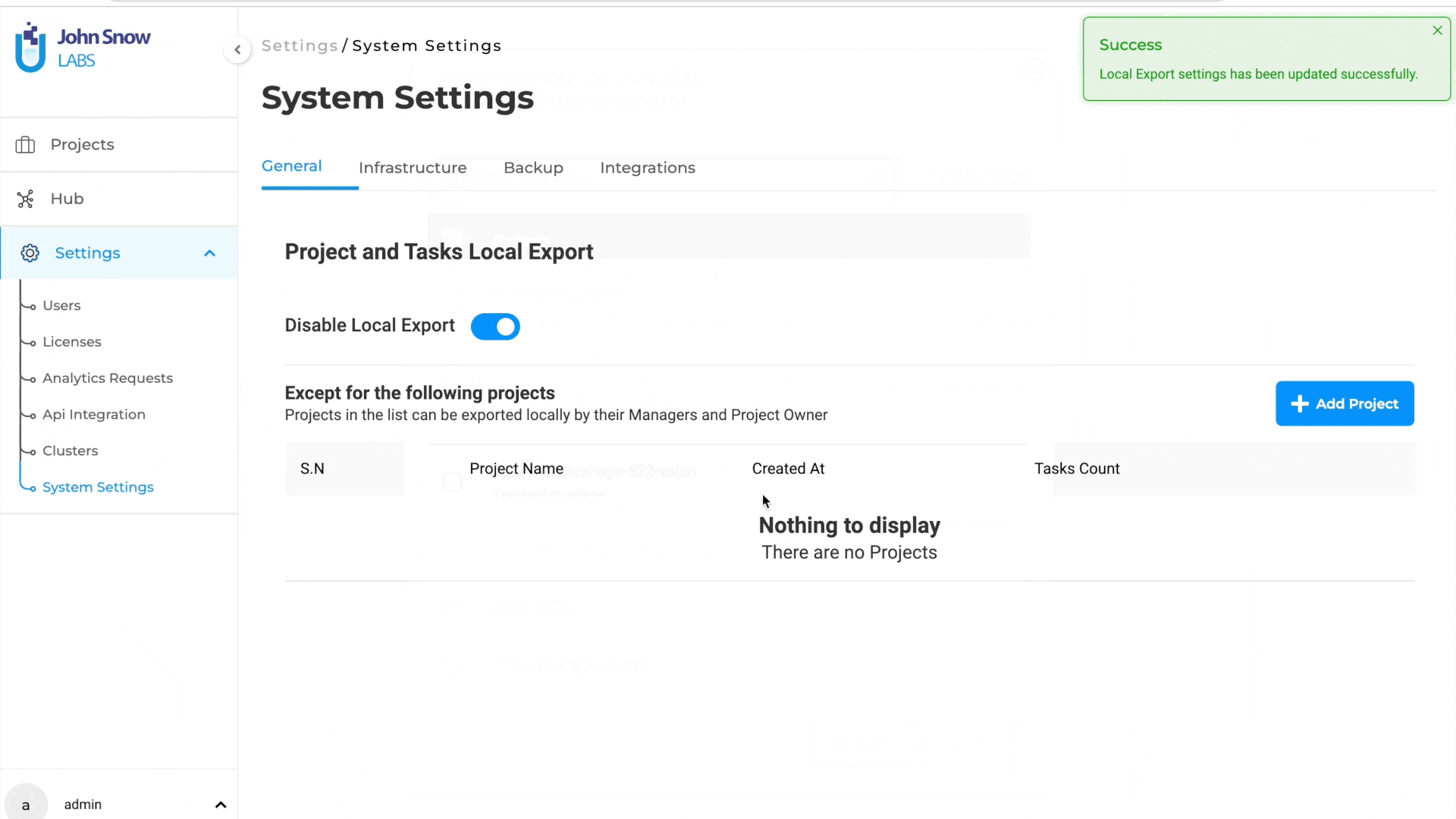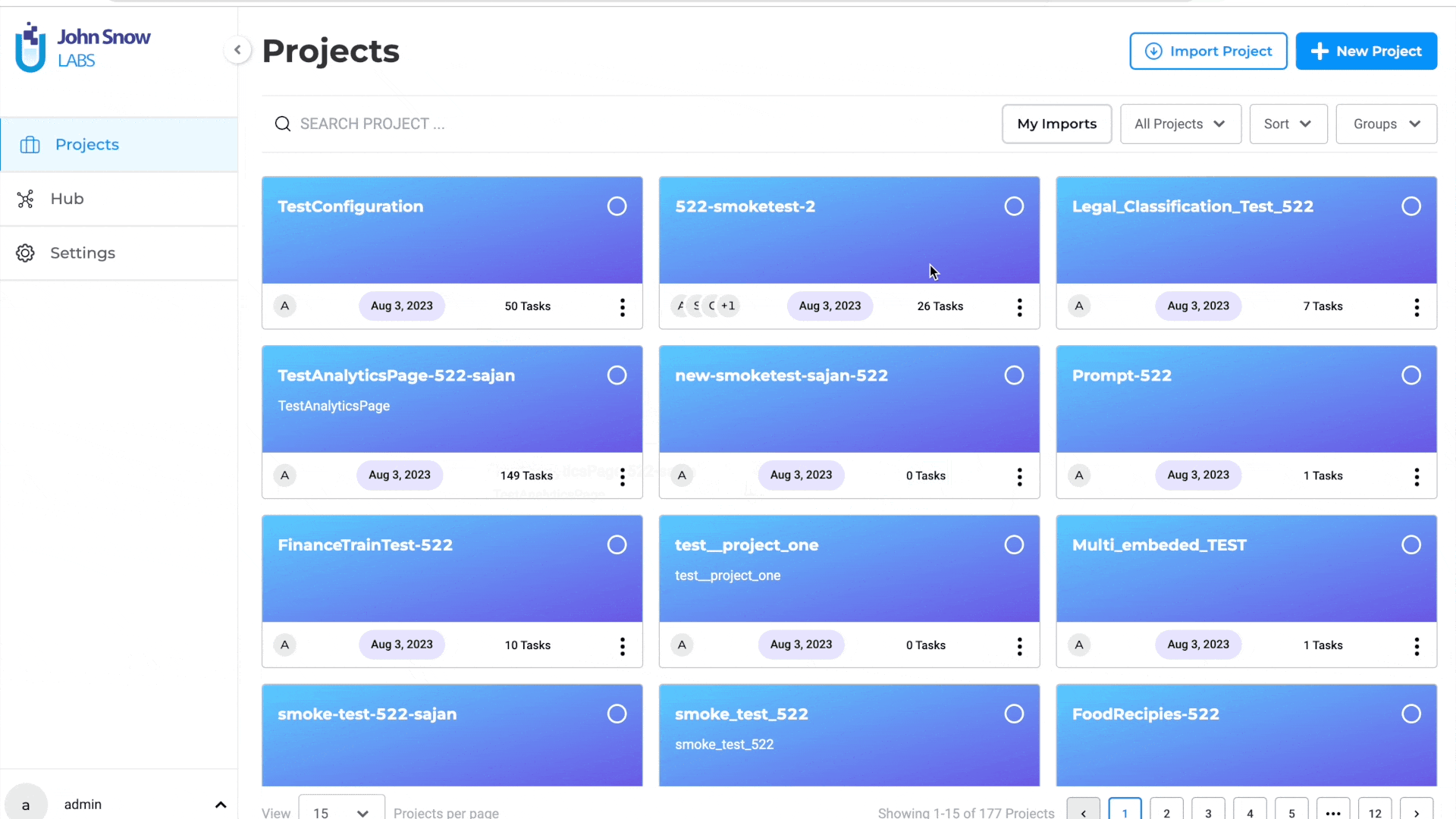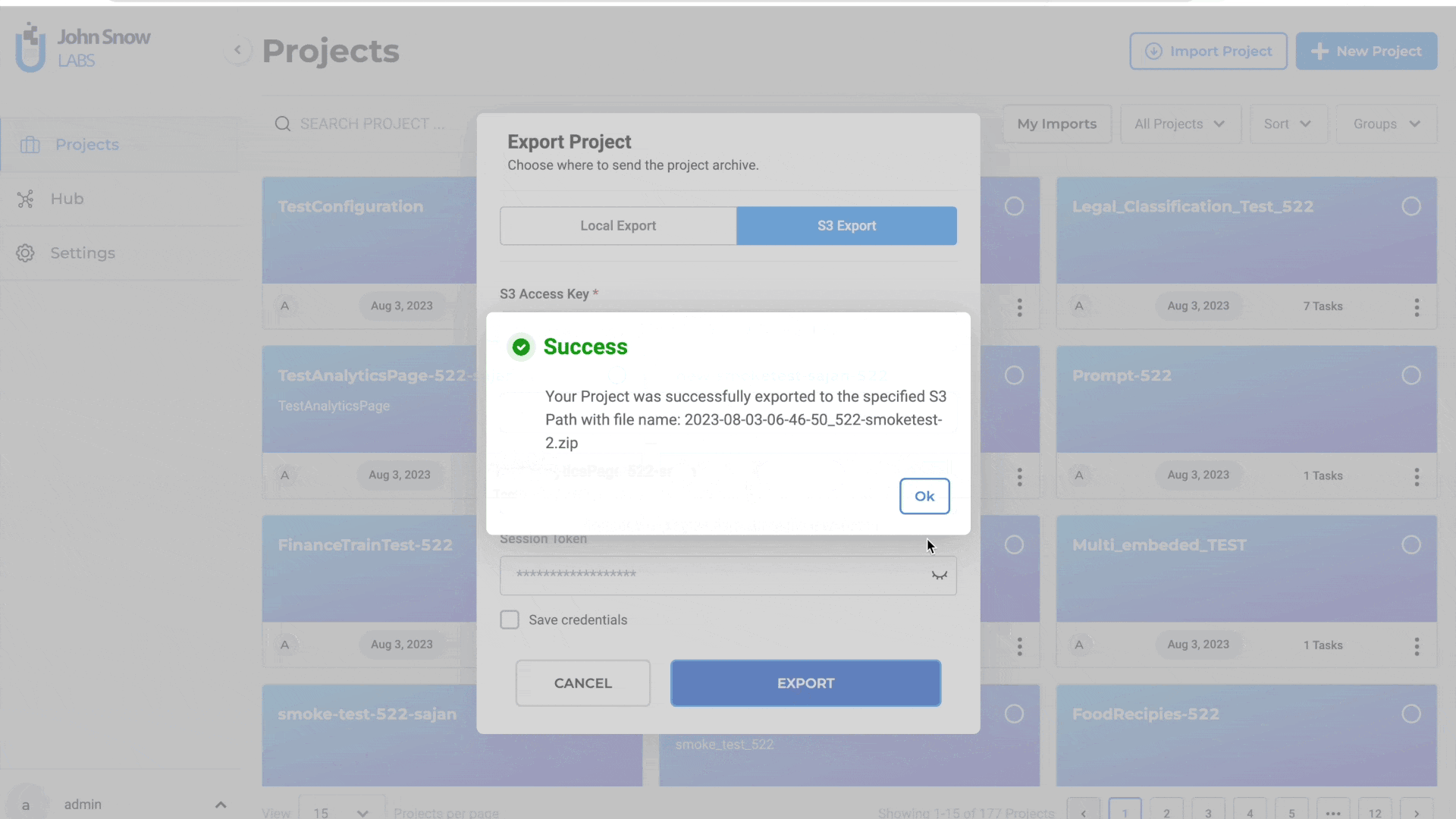
Improved HIPAA Compliance with Restricted Local Exports
Administrators can restrict local exports to enhance data security and maintain HIPAA compliance.
When local export is disabled, users can no longer download projects or tasks to their workstation and must instead export them securely to cloud storage, such as Amazon S3 or Azure Blob.
This ensures sensitive information remains protected while still allowing flexible and compliant data handling for annotation workflows.
Disable Local Export:
System administrators can now manage export settings from the system settings page. By enabling the “Disable Local Export” option, the export to a local workstation for all projects is turned off.
Selective Export Exceptions:
Administrators have the flexibility to specify projects that can still use local export if needed. To do this, click on the “Add Project” button from the Exceptions widget and search for the projects to add to the exceptions list.
S3 Bucket Export:
With the “Disable Local Export” option activated, users can only export tasks and projects to Amazon S3 bucket paths. This ensures the protection of sensitive data that will be stored securely in the cloud.