Description
The GeoLayoutLM model, introduced in the paper “GeoLayoutLM: Geometric Pre-training for Visual Information Extraction” by Chuwei Luo, Changxu Cheng, Qi Zheng, and Cong Yao, presents a new approach for visual information extraction (VIE) using geometric pre-training.
GeoLayoutLM is designed as a multi-modal framework that handles tasks like Semantic Entity Recognition (SER) and Relation Extraction (RE). What sets it apart is the use of geometric pre-training, along with specialized relation heads that are fine-tuned for the RE task. These features help to improve the model’s understanding of spatial relationships in documents. As a result, GeoLayoutLM achieves strong performance in SER and significantly outperforms previous models in RE, showcasing its ability to handle specific tasks in the VIE field.
Predicted Entities
other, b-header, i-header, b-question, i-question, b-answer, i-answer.
Live Demo Open in Colab Download –> Copy S3 URI
How to use
binary_to_image = BinaryToImage() \
.setImageType(ImageType.TYPE_3BYTE_BGR)
ocr = ImageToHocr() \
.setInputCol("image") \
.setOutputCol("hocr") \
.setIgnoreResolution(False) \
.setOcrParams(["preserve_interword_spaces=0"]) \
.setPageSegMode(PageSegmentationMode.SPARSE_TEXT)
tokenizer = BrosHocrTokenizer.pretrained("bros_hocr_tokenizer", "en", "clinical/ocr/") \
.setInputCol("hocr") \
.setOutputCol("tokens")
hocr_to_features = HocrToFeatures() \
.setInputCols(["tokens", "image"]) \
.setOutputCol("features")
ner = VisualDocumentNerGeo().pretrained("visual_ner_geo_v1", "en", "clinical/ocr/") \
.setInputCols(["features", "tokens", "image"]) \
.setWhiteList(["other", "i-header", "b-header", "i-question", "b-question", "i-answer", "b-answer"]) \
.setLabels(["other", "i-header", "b-header", "i-question", "b-question", "i-answer", "b-answer"]) \
.setOutputCol("entities")
draw_annotations = ImageDrawAnnotations() \
.setInputCol("image") \
.setInputChunksCol("entities") \
.setOutputCol("image_with_annotations") \
.setFilledRect(False)
hocr_to_features1 = HocrToFeatures() \
.setInputCols(["entities", "image"]) \
.setOutputCol("features1")
re = GeoRelationExtractor.pretrained("visual_re_geo_v2", "en", "clinical/ocr/") \
.setInputCols(("features1", "entities", "image")) \
.setLabels(( "other", "b-header", "i-header", "b-question", "b-question", "b-answer", "i-answer")) \
.setOutputCol("relations") \
.setOutputFormat(RelationOutputFormat.ANNOTATIONS)
pipeline = PipelineModel(stages=[
binary_to_image,
ocr,
tokenizer,
hocr_to_features,
ner,
hocr_to_features1,
re,
draw_annotations
])
img_path = '/content/imgs/'
img_example_df = spark.read.format("binaryFile").load(img_path).cache()
result = pipeline.transform(img_example_df)
val binary_to_image = new BinaryToImage()
.setImageType(ImageType.TYPE_3BYTE_BGR)
val ocr = new ImageToHocr()
.setInputCol("image")
.setOutputCol("hocr")
.setIgnoreResolution(False)
.setOcrParams(Array("preserve_interword_spaces=0"))
.setPageSegMode(PageSegmentationMode.SPARSE_TEXT)
val tokenizer = new BrosHocrTokenizer.pretrained("bros_hocr_tokenizer", "en", "clinical/ocr/")
.setInputCol("hocr")
.setOutputCol("tokens")
val hocr_to_features = new HocrToFeatures()
.setInputCols(Array("tokens", "image"))
.setOutputCol("features")
val ner = new VisualDocumentNerGeo().pretrained("visual_ner_geo_v1", "en", "clinical/ocr/")
.setInputCols(Array("features", "tokens", "image"))
.setWhiteList(Array("other", "i-header", "b-header", "i-question", "b-question", "i-answer", "b-answer"))
.setLabels(Array("other", "i-header", "b-header", "i-question", "b-question", "i-answer", "b-answer"))
.setOutputCol("entities")
val draw_annotations = new ImageDrawAnnotations()
.setInputCol("image")
.setInputChunksCol("entities")
.setOutputCol("image_with_annotations")
.setFilledRect(False)
val hocr_to_features1 = new HocrToFeatures()
.setInputCols(Array("entities", "image"))
.setOutputCol("features1")
val re = new GeoRelationExtractor.pretrained("visual_re_geo_v2", "en", "clinical/ocr/")
.setInputCols(("features1", "entities", "image"))
.setLabels(("other", "b-header", "i-header", "b-question", "b-question", "b-answer", "i-answer"))
.setOutputCol("relations")
.setOutputFormat(RelationOutputFormat.ANNOTATIONS)
val pipeline = new PipelineModel().setStages(Array(
binary_to_image,
ocr,
tokenizer,
hocr_to_features,
ner,
hocr_to_features1,
re,
draw_annotations))
val img_path = "/content/imgs/"
val img_example_df = spark.read.format("binaryFile").load(img_path).cache()
val result = pipeline.transform(img_example_df)
Example
Input:

Output:
Image with annotations:
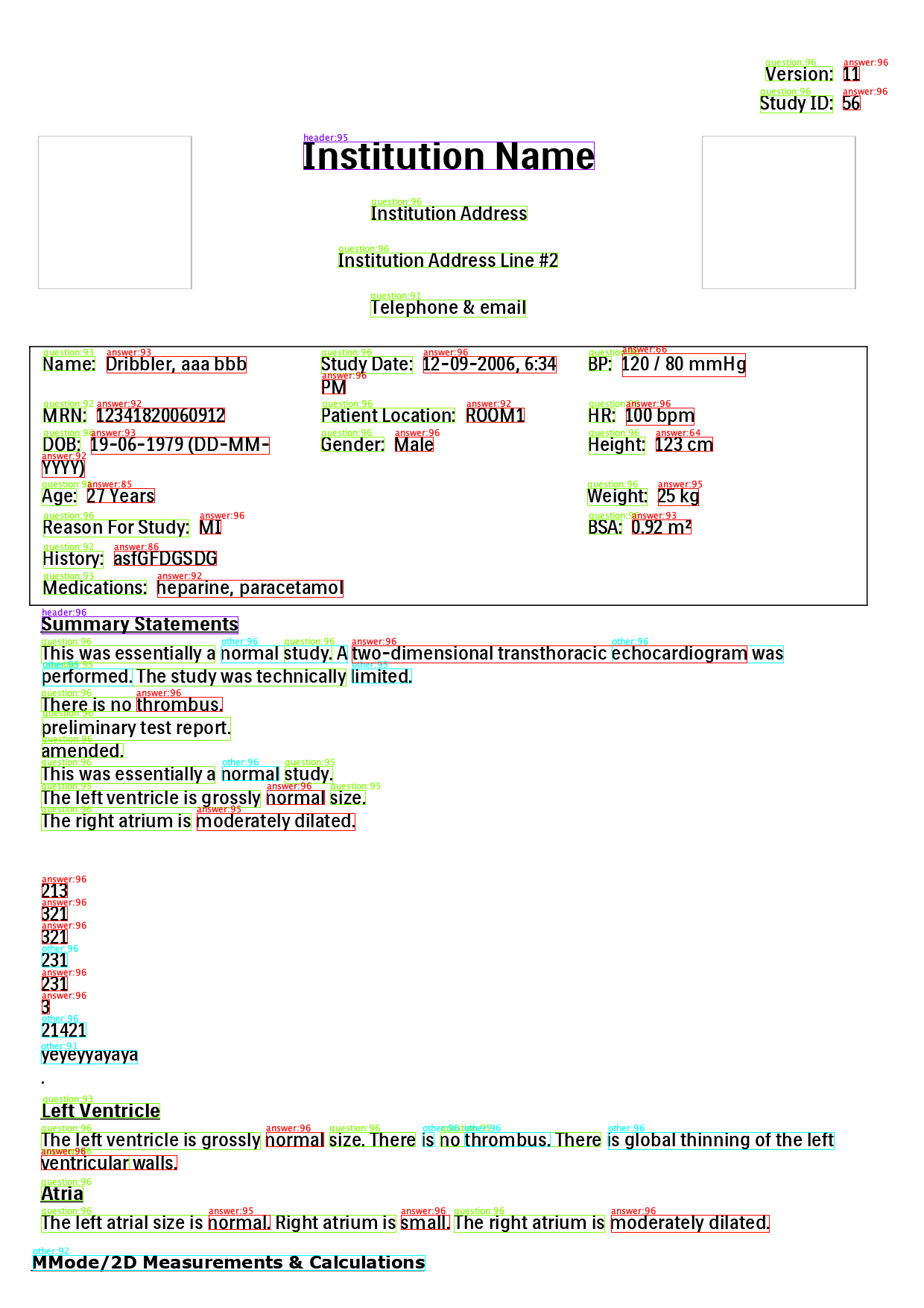
Result:
+-------------------------------+-----------------+--------------+------------------+--------------+
|result |text1 |bbox1 |text2 |bbox2 |
+-------------------------------+-----------------+--------------+------------------+--------------+
|Version: -> 11 |Version: |1027 89 90 19 |11 |1132 89 21 19 |
|Study ID: -> 56 |Study ID: |1020 128 97 23|56 |1131 128 23 19|
|Name: -> Dribbler, aaa bbb |Name: |58 478 69 19 |Dribbler, aaa bbb |143 478 187 22|
|Study Date: -> 12-09-2006, 6:34|Study Date: |431 478 122 23|12-09-2006, 6:34 |568 478 178 22|
|BP: -> 120 / 80 mmHg |BP: |790 478 30 19 |120 / 80 mmHg |835 474 165 31|
|MRN: -> 12341820060912 |MRN: |58 547 57 19 |12341820060912 |130 547 171 19|
|Patient Location: -> ROOM1 |Patient Location:|432 547 178 19|ROOM1 |626 547 77 19 |
|Patient Location: -> 100 bpm |Patient Location:|432 547 178 19|100 bpm |840 547 91 23 |
|DOB: -> 19-06-1979 (DD-MM- |DOB: |58 586 49 19 |19-06-1979 (DD-MM-|122 586 239 23|
|Gender: -> Male |Gender: |431 586 84 19 |Male |530 586 51 19 |
|Height: -> 123 cm |Height: |790 586 75 23 |123 cm |880 586 76 19 |
|Age: -> 27 Years |Age: |56 655 46 23 |27 Years |117 655 90 19 |
|Reason For Study: -> MI |Reason For Study:|58 697 195 23 |MI |268 697 28 19 |
+-------------------------------+-----------------+--------------+------------------+--------------+
Model Information
| Model Name: | visual_re_geo_v2 |
| Type: | ocr |
| Compatibility: | Visual NLP 5.4.1+ |
| License: | Licensed |
| Edition: | Official |
| Language: | en |