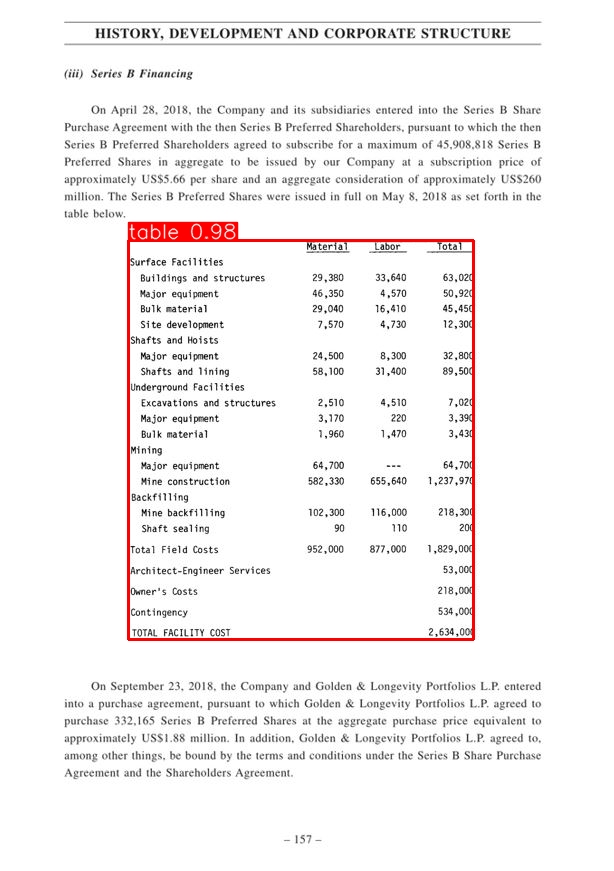
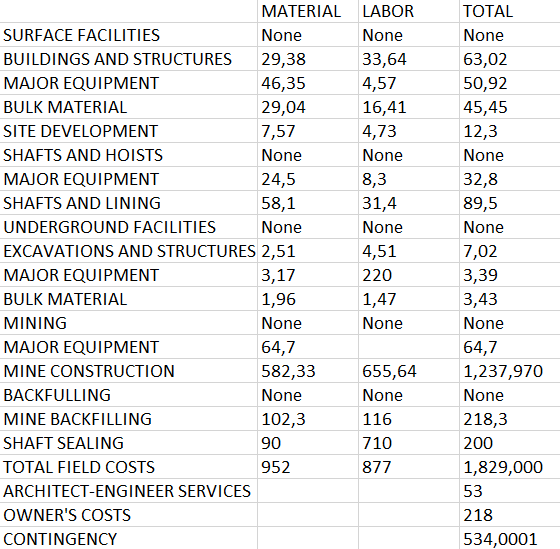
Description
This model shows the capabilities for table recognition and free-text extraction using OCR techniques. For table recognition is proposed a CascadTabNet model. CascadeTabNet is a machine learning model for table detection in document images. It is based on a cascaded architecture, which is a two-stage process where the model first detects candidate regions that may contain tables, and then classifies these regions as tables or non-tables. The model is trained using a dataset of document images, where the tables have been manually annotated.
The benchmark results show that the model is able to detect tables in document images with high accuracy. On the ICDAR2013 table competition dataset, CascadeTabNet achieved an F1-score of 0.85, which is considered a good score in this dataset. On the COCO-Text dataset, the model achieved a precision of 0.82 and a recall of 0.79, which are also considered good scores. In addition, the model has been evaluated on the UNLV dataset, where it achieved a precision of 0.87 and a recall of 0.83.
Predicted Entities
How to use
binary_to_image = BinaryToImage() \
.setInputCol("content") \
.setOutputCol("image") \
.setImageType(ImageType.TYPE_3BYTE_BGR)
# Detect tables on the page using pretrained model
# It can be finetuned for have more accurate results for more specific documents
table_detector = ImageTableDetector.pretrained("general_model_table_detection_v2", "en", "clinical/ocr") \
.setInputCol("image") \
.setOutputCol("region")
# Draw detected region's with table to the page
draw_regions = ImageDrawRegions() \
.setInputCol("image") \
.setInputRegionsCol("region") \
.setOutputCol("image_with_regions") \
.setRectColor(Color.red)
# Extract table regions to separate images
splitter = ImageSplitRegions() \
.setInputCol("image") \
.setInputRegionsCol("region") \
.setOutputCol("table_image") \
.setDropCols("image")
# Detect cells on the table image
cell_detector = ImageTableCellDetector() \
.setInputCol("table_image") \
.setOutputCol("cells") \
.setAlgoType("morphops") \
.setDrawDetectedLines(True)
# Extract text from the detected cells
table_recognition = ImageCellsToTextTable() \
.setInputCol("table_image") \
.setCellsCol('cells') \
.setMargin(3) \
.setStrip(True) \
.setOutputCol('table')
# Erase detected table regions
fill_regions = ImageDrawRegions() \
.setInputCol("image") \
.setInputRegionsCol("region") \
.setOutputCol("image_1") \
.setRectColor(Color.white) \
.setFilledRect(True)
# OCR
ocr = ImageToText() \
.setInputCol("image_1") \
.setOutputCol("text") \
.setOcrParams(["preserve_interword_spaces=1", ]) \
.setKeepLayout(True) \
.setOutputSpaceCharacterWidth(8)
pipeline_table = PipelineModel(stages=[
binary_to_image,
table_detector,
draw_regions,
fill_regions,
splitter,
cell_detector,
table_recognition,
ocr
])
imagePath = "/content/cTDaR_t10096.jpg"
df = spark.read.format("binaryFile").load(imagePath)
tables_results = pipeline_table.transform(df).cache()
val binary_to_image = new BinaryToImage()
.setInputCol("content")
.setOutputCol("image")
.setImageType(ImageType.TYPE_3BYTE_BGR)
# Detect tables on the page using pretrained model
# It can be finetuned for have more accurate results for more specific documents
val table_detector = ImageTableDetector
.pretrained("general_model_table_detection_v2", "en", "clinical/ocr")
.setInputCol("image")
.setOutputCol("region")
# Draw detected region's with table to the page
val draw_regions = new ImageDrawRegions()
.setInputCol("image")
.setInputRegionsCol("region")
.setOutputCol("image_with_regions")
.setRectColor(Color.red)
# Extract table regions to separate images
val splitter = new ImageSplitRegions()
.setInputCol("image")
.setInputRegionsCol("region")
.setOutputCol("table_image")
.setDropCols("image")
# Detect cells on the table image
val cell_detector = new ImageTableCellDetector()
.setInputCol("table_image")
.setOutputCol("cells")
.setAlgoType("morphops")
.setDrawDetectedLines(True)
# Extract text from the detected cells
val table_recognition = new ImageCellsToTextTable()
.setInputCol("table_image")
.setCellsCol("cells")
.setMargin(3)
.setStrip(True)
.setOutputCol("table")
# Erase detected table regions
val fill_regions = new ImageDrawRegions()
.setInputCol("image")
.setInputRegionsCol("region")
.setOutputCol("image_1")
.setRectColor(Color.white)
.setFilledRect(True)
# OCR
val ocr = new ImageToText()
.setInputCol("image_1")
.setOutputCol("text")
.setOcrParams(Array("preserve_interword_spaces=1", ))
.setKeepLayout(True)
.setOutputSpaceCharacterWidth(8)
val pipeline_table = new PipelineModel().setStages(Array(
binary_to_image,
table_detector,
draw_regions,
fill_regions,
splitter,
cell_detector,
table_recognition,
ocr))
val imagePath = "/content/cTDaR_t10096.jpg"
val df = spark.read.format("binaryFile").load(imagePath)
val tables_results = pipeline_table.transform(df).cache()
Example
Input Image

Table Structure Recognition
Model Information
| Model Name: | table_recognition |
| Compatibility: | Healthcare NLP 4.1.0+ |
| License: | Licensed |
| Edition: | Official |
| Language: | en |